Gene
KWMTBOMO03936 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010231
Annotation
cuticular_protein_RR-2_motif_68_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.493 PlasmaMembrane Reliability : 1.047
Sequence
CDS
ATGGCCCTCAAGTTGGTAACCTTTTCTTGCCTTTTGGCTGCCGCTTATGGGAGCGTTGTTCCCGCCGCCTATGCAGCTGCGCCTCTGGTAGCAGCAGCTCCTGCAGTAGCGGCAAGAATCGAAGAATTCGATCCTCTTCCTCAATACAGATTTGGCTATGATGTAGCGGACTCTTTAACCGGTGATTACAAAAGTCAACAAGAAGAACGCAATGGAGATTTGGTTCAAGGATCTTACTCTTTGGTTGAACCCGACGGCACACGTCGTGTCGTGGACTACGCCGCTGACTCCATTAATGGCTTCAATGCCGTAGTACGTAAAGAGCCACTCGTGGCGGCTGCGCCTGCAGTTGTTGCAGAGCCCGCAGTAGTACCCGCTCGTTATGCAGCAGCTCCGGTGGCTGCTGCTCCTGTAGCCGCTGCTCCTATAGCCGCTGCCCCTGTTGCAGCACCGTACTACGCCGCTCGCTACGCAGTTGCTCCTGCATATTCGGCACCCTATGTTACTGCAGCCAGATATGAAGCCGCTCCCTTCGTAGCACGGTACGCTGCTGCTCCAGTAGCTGCTGCACCGGTCGCTGCTGCACCGGCTCCTATCTACGCTCGTTATGCGACCGCACCAGTAGCTCCCGCACCTGTTGTTGCTGCGCGCTACGCAGCTCCTTCCGTAGTTGCAGCACGCTATGCTGCAGCTGGCCCATATGCTGCCTACACAGCTCCATTTTCTGCAGCCTACACAGCTCCATTTTCTGCGGCCTACACCTCATATGCCTCTCCAGTAGCTGCAGCATACACCACCTACACCGCAACGGGACCCGTGTCAGCAGCTCCAGTTGCTGCCGCGTACGCTGCTCCTATCGCTGCGACAGCGGTACCAGCTGTGGCCTCCACCGTTCGTGCAGCTCCCGCTAAGCTCATTGAAACTACAGCTGCTGGATACCGTTACCCCTAA
Protein
MALKLVTFSCLLAAAYGSVVPAAYAAAPLVAAAPAVAARIEEFDPLPQYRFGYDVADSLTGDYKSQQEERNGDLVQGSYSLVEPDGTRRVVDYAADSINGFNAVVRKEPLVAAAPAVVAEPAVVPARYAAAPVAAAPVAAAPIAAAPVAAPYYAARYAVAPAYSAPYVTAARYEAAPFVARYAAAPVAAAPVAAAPAPIYARYATAPVAPAPVVAARYAAPSVVAARYAAAGPYAAYTAPFSAAYTAPFSAAYTSYASPVAAAYTTYTATGPVSAAPVAAAYAAPIAATAVPAVASTVRAAPAKLIETTAAGYRYP
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
Topology
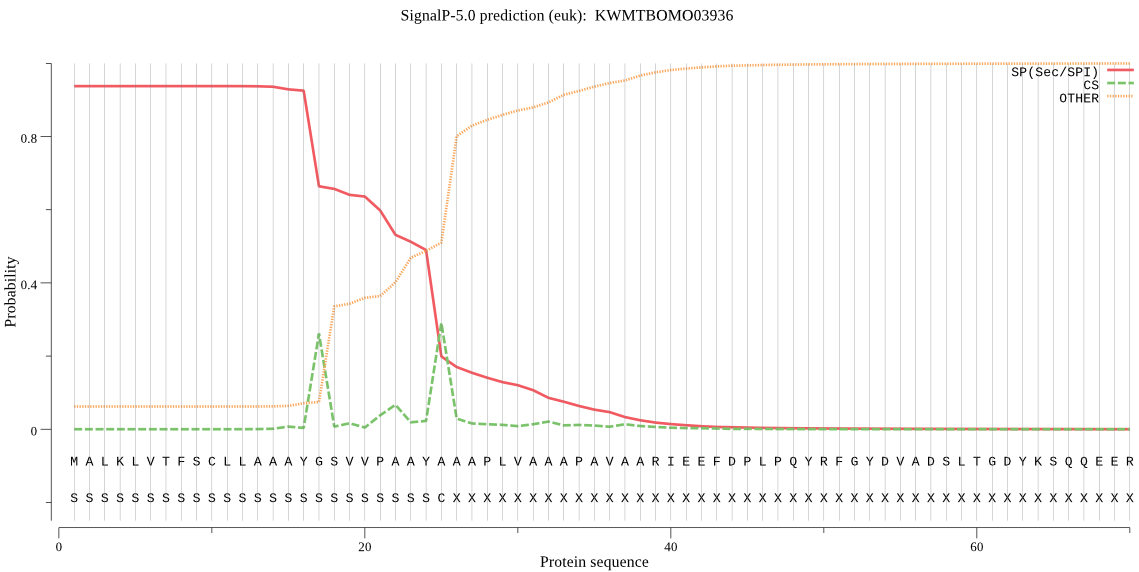
SignalP
Position: 1 - 25,
Likelihood: 0.937804
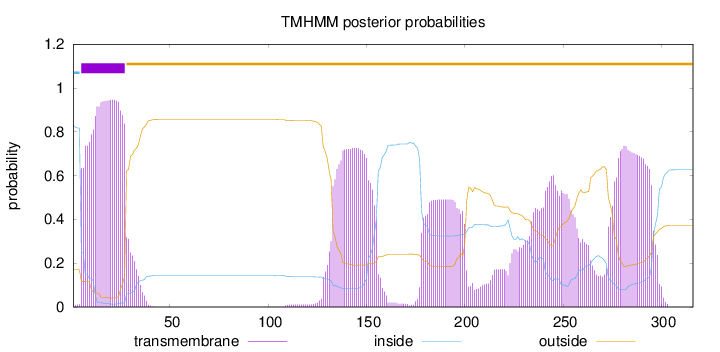
Length:
316
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
86.05316
Exp number, first 60 AAs:
21.75938
Total prob of N-in:
0.82905
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 316
Population Genetic Test Statistics
Pi
312.970608
Theta
204.888167
Tajima's D
1.640393
CLR
0.016655
CSRT
0.816359182040898
Interpretation
Uncertain
