Gene
KWMTBOMO03935 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010145
Annotation
cuticular_protein_RR-2_motif_67_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.585
Sequence
CDS
ATGGCATTCAAGTTCGTAGTATTGGCTTGCCTGGTAGCTGTTGCCAGCGCCGGTGTGGTTCCAGTAGCGCAATACGGTTACGCAGCACCCGCATTGCACGCAGCACCAGTGTCGTACTCCGCCCCCATTGCTAAAGTCGCTGTCGAAGAATACGATGCACACCCTCAATACAGTTTCGCATACGACGTACAAGATGGCGTCACTGGTGACTCGAAGAGTCAACACGAGACCCGCGACGGCGATGTCGTTCAAGGCTCTTACTCCGTGGTTGACCCCGATGGCATCAAGCGCACCGTTGAATACACCGCCGATCCCCACAACGGCTTCAACGCTGTCGTGCACAGAGAACCCCTCGGCCACGCCGCTAAAGTAGCATATGCTGCACCAGTCGCCAAGATTGCTGCCCCAGTTACTTATGCCGCATCCCCGGTCGTACACTCTGCTCCTATCGTGCACTCCGCCCCTGTCGCCTACTCATCCCCTATCGCTAAATACCCTGCCCCCTTCACCTATTCCGCTCCCATCTACCACCACTAA
Protein
MAFKFVVLACLVAVASAGVVPVAQYGYAAPALHAAPVSYSAPIAKVAVEEYDAHPQYSFAYDVQDGVTGDSKSQHETRDGDVVQGSYSVVDPDGIKRTVEYTADPHNGFNAVVHREPLGHAAKVAYAAPVAKIAAPVTYAASPVVHSAPIVHSAPVAYSSPIAKYPAPFTYSAPIYHH
Summary
Uniprot
C0H6Q9
H9JKU4
A0A194QZR3
A0A194PLK4
I4DKC3
C0H6R3
+ More
I4DMZ9 A0A2W1BX58 A0A2A4K2Q5 B2DBJ1 A0A3S2LNF3 B2DBK4 A0A194PN94 A0A194QZS3 B2DBJ2 A0A194PMK1 A0A2W1BQU7 I4DJK8 I4DKC4 A0A2A4K0Z2 I4DN00 A0A194PLA1 I4DJV8 A0A194PMJ2 A0A2H1VZN6 A0A194R0K2 A0A194PSV5 A0A0R7JPI3 A0A194QZ91 A0A2A4K1L9 A0A2A4K203 A0A194QZ16 A0A2H1W045 A0A3S2M3H7 Q9BPR3 A0A2A4K2R9 A0A194R4J0 A0A3Q8HNV7 A0A2W1BQM3 A0A3Q8HG34 A0A084WBR7 A0A084WBR6 A0A084WBR8 A0A3Q8HGL9 A0A182FQD1 A0A182VZH1 A0A182LXP9 A0A2H1VZQ5 A0A182PGI9 A0A212EVZ4 A0A182VZG9 A0A3F2YUZ3 Q380K3 A0A182HLH6 A0A182JJP4 D6W8Q5 D6W8R0 D6W8Q8 A0A182WSM6 A0A0L7QWA1 N6TQ67 N6TX66 A0A3Q8HG43 A0A2H1VZN4 K7IX05 A0A0A1XB19 A0A154P833 A0A034VMZ8 W5JVR9
I4DMZ9 A0A2W1BX58 A0A2A4K2Q5 B2DBJ1 A0A3S2LNF3 B2DBK4 A0A194PN94 A0A194QZS3 B2DBJ2 A0A194PMK1 A0A2W1BQU7 I4DJK8 I4DKC4 A0A2A4K0Z2 I4DN00 A0A194PLA1 I4DJV8 A0A194PMJ2 A0A2H1VZN6 A0A194R0K2 A0A194PSV5 A0A0R7JPI3 A0A194QZ91 A0A2A4K1L9 A0A2A4K203 A0A194QZ16 A0A2H1W045 A0A3S2M3H7 Q9BPR3 A0A2A4K2R9 A0A194R4J0 A0A3Q8HNV7 A0A2W1BQM3 A0A3Q8HG34 A0A084WBR7 A0A084WBR6 A0A084WBR8 A0A3Q8HGL9 A0A182FQD1 A0A182VZH1 A0A182LXP9 A0A2H1VZQ5 A0A182PGI9 A0A212EVZ4 A0A182VZG9 A0A3F2YUZ3 Q380K3 A0A182HLH6 A0A182JJP4 D6W8Q5 D6W8R0 D6W8Q8 A0A182WSM6 A0A0L7QWA1 N6TQ67 N6TX66 A0A3Q8HG43 A0A2H1VZN4 K7IX05 A0A0A1XB19 A0A154P833 A0A034VMZ8 W5JVR9
Pubmed
EMBL
BR000568
FAA00570.1
BABH01011539
KQ460930
KPJ10774.1
KQ459600
+ More
KPI94207.1 AK401741 BAM18363.1 BABH01011540 BR000572 FAA00574.1 AK402667 BAM19289.1 KZ149930 PZC77400.1 NWSH01000258 PCG77960.1 AB264671 BAG30750.1 RSAL01000049 RVE50420.1 AB264688 BAG30763.1 KPI94209.1 KPJ10784.1 AB264674 BAG30751.1 KPI94218.1 PZC77398.1 AK401476 BAM18098.1 AK401742 BAM18364.1 PCG77945.1 AK402668 BAM19290.1 KPI94211.1 AK401576 BAM18198.1 KPI94208.1 ODYU01005440 SOQ46301.1 KPJ10775.1 KPI94210.1 KP072002 AKQ00283.1 KPJ10777.1 PCG77959.1 PCG77958.1 KPJ10778.1 SOQ46296.1 RVE50423.1 AB047484 BAB32481.1 PCG77942.1 KPJ10776.1 MG601638 AYA49963.1 PZC77402.1 MG601639 AYA49964.1 ATLV01022427 KE525332 KFB47661.1 ATLV01022426 KFB47660.1 KFB47662.1 MG601698 AYA50023.1 AXCM01013007 SOQ46298.1 AGBW02012104 OWR45665.1 AAAB01008987 EAL38559.1 APCN01000901 AXCP01009407 KQ971312 EEZ98282.1 EEZ98278.1 EEZ98280.1 KQ414716 KOC62887.1 APGK01011108 KB738393 ENN82624.1 APGK01011109 KB738394 ENN82623.1 MG601642 AYA49967.1 SOQ46299.1 AAZX01011882 GBXI01006332 JAD07960.1 KQ434823 KZC07280.1 GAKP01015450 JAC43502.1 ADMH02000274 ETN67160.1
KPI94207.1 AK401741 BAM18363.1 BABH01011540 BR000572 FAA00574.1 AK402667 BAM19289.1 KZ149930 PZC77400.1 NWSH01000258 PCG77960.1 AB264671 BAG30750.1 RSAL01000049 RVE50420.1 AB264688 BAG30763.1 KPI94209.1 KPJ10784.1 AB264674 BAG30751.1 KPI94218.1 PZC77398.1 AK401476 BAM18098.1 AK401742 BAM18364.1 PCG77945.1 AK402668 BAM19290.1 KPI94211.1 AK401576 BAM18198.1 KPI94208.1 ODYU01005440 SOQ46301.1 KPJ10775.1 KPI94210.1 KP072002 AKQ00283.1 KPJ10777.1 PCG77959.1 PCG77958.1 KPJ10778.1 SOQ46296.1 RVE50423.1 AB047484 BAB32481.1 PCG77942.1 KPJ10776.1 MG601638 AYA49963.1 PZC77402.1 MG601639 AYA49964.1 ATLV01022427 KE525332 KFB47661.1 ATLV01022426 KFB47660.1 KFB47662.1 MG601698 AYA50023.1 AXCM01013007 SOQ46298.1 AGBW02012104 OWR45665.1 AAAB01008987 EAL38559.1 APCN01000901 AXCP01009407 KQ971312 EEZ98282.1 EEZ98278.1 EEZ98280.1 KQ414716 KOC62887.1 APGK01011108 KB738393 ENN82624.1 APGK01011109 KB738394 ENN82623.1 MG601642 AYA49967.1 SOQ46299.1 AAZX01011882 GBXI01006332 JAD07960.1 KQ434823 KZC07280.1 GAKP01015450 JAC43502.1 ADMH02000274 ETN67160.1
Proteomes
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6Q9
H9JKU4
A0A194QZR3
A0A194PLK4
I4DKC3
C0H6R3
+ More
I4DMZ9 A0A2W1BX58 A0A2A4K2Q5 B2DBJ1 A0A3S2LNF3 B2DBK4 A0A194PN94 A0A194QZS3 B2DBJ2 A0A194PMK1 A0A2W1BQU7 I4DJK8 I4DKC4 A0A2A4K0Z2 I4DN00 A0A194PLA1 I4DJV8 A0A194PMJ2 A0A2H1VZN6 A0A194R0K2 A0A194PSV5 A0A0R7JPI3 A0A194QZ91 A0A2A4K1L9 A0A2A4K203 A0A194QZ16 A0A2H1W045 A0A3S2M3H7 Q9BPR3 A0A2A4K2R9 A0A194R4J0 A0A3Q8HNV7 A0A2W1BQM3 A0A3Q8HG34 A0A084WBR7 A0A084WBR6 A0A084WBR8 A0A3Q8HGL9 A0A182FQD1 A0A182VZH1 A0A182LXP9 A0A2H1VZQ5 A0A182PGI9 A0A212EVZ4 A0A182VZG9 A0A3F2YUZ3 Q380K3 A0A182HLH6 A0A182JJP4 D6W8Q5 D6W8R0 D6W8Q8 A0A182WSM6 A0A0L7QWA1 N6TQ67 N6TX66 A0A3Q8HG43 A0A2H1VZN4 K7IX05 A0A0A1XB19 A0A154P833 A0A034VMZ8 W5JVR9
I4DMZ9 A0A2W1BX58 A0A2A4K2Q5 B2DBJ1 A0A3S2LNF3 B2DBK4 A0A194PN94 A0A194QZS3 B2DBJ2 A0A194PMK1 A0A2W1BQU7 I4DJK8 I4DKC4 A0A2A4K0Z2 I4DN00 A0A194PLA1 I4DJV8 A0A194PMJ2 A0A2H1VZN6 A0A194R0K2 A0A194PSV5 A0A0R7JPI3 A0A194QZ91 A0A2A4K1L9 A0A2A4K203 A0A194QZ16 A0A2H1W045 A0A3S2M3H7 Q9BPR3 A0A2A4K2R9 A0A194R4J0 A0A3Q8HNV7 A0A2W1BQM3 A0A3Q8HG34 A0A084WBR7 A0A084WBR6 A0A084WBR8 A0A3Q8HGL9 A0A182FQD1 A0A182VZH1 A0A182LXP9 A0A2H1VZQ5 A0A182PGI9 A0A212EVZ4 A0A182VZG9 A0A3F2YUZ3 Q380K3 A0A182HLH6 A0A182JJP4 D6W8Q5 D6W8R0 D6W8Q8 A0A182WSM6 A0A0L7QWA1 N6TQ67 N6TX66 A0A3Q8HG43 A0A2H1VZN4 K7IX05 A0A0A1XB19 A0A154P833 A0A034VMZ8 W5JVR9
Ontologies
GO
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.990011
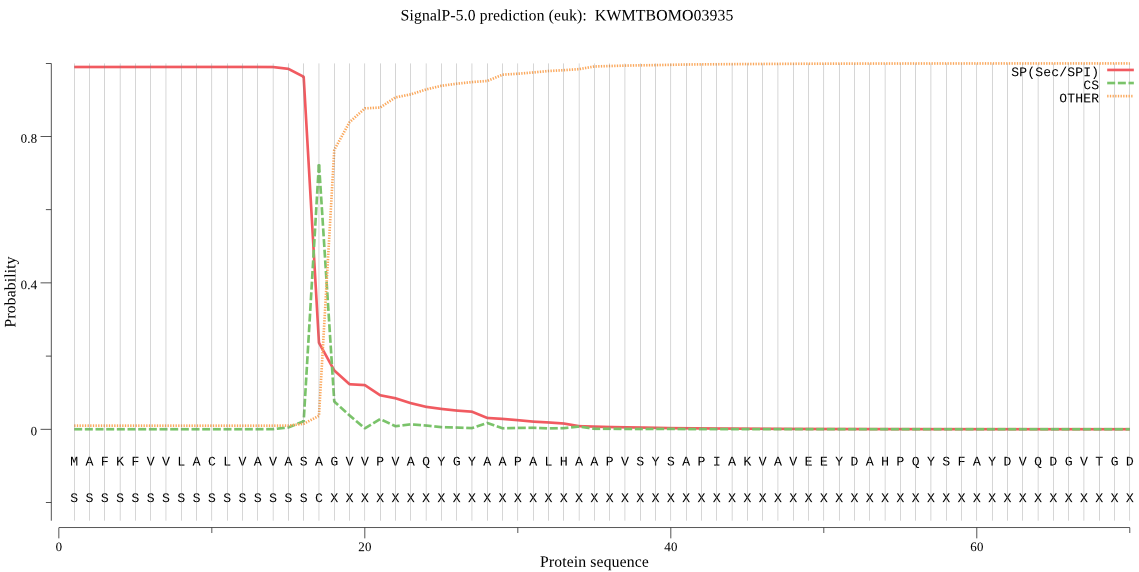
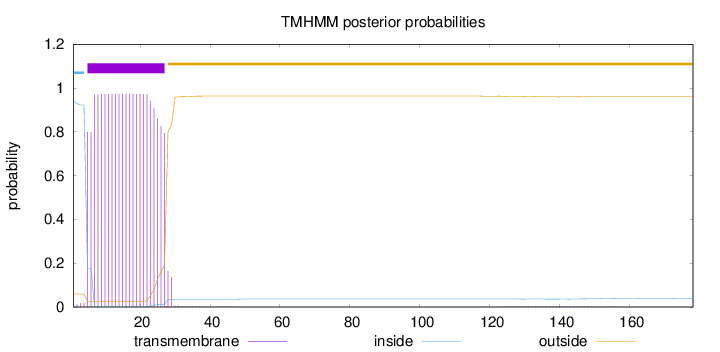
Length:
178
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.99532
Exp number, first 60 AAs:
21.88824
Total prob of N-in:
0.94064
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 178
Population Genetic Test Statistics
Pi
215.09773
Theta
140.224344
Tajima's D
0.719023
CLR
0.342676
CSRT
0.577221138943053
Interpretation
Uncertain
