Gene
KWMTBOMO03926
Pre Gene Modal
BGIBMGA010150
Annotation
Uncharacterized_protein_OBRU01_03101_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.297
Sequence
CDS
ATGGGACAAAGTTCTGTGAGTACAACCAAATTACCCACTGAAAGTAGTCCCGATTTTTCTGCTTCAATAATGTGTAAAACAATTGAAGAAGATGGAATGACGAAACTAAAATTGTACCAAAACAAAGAAAACACTATAGTTCCACTACATTGGTTTGGTGTTTTGGTGCCGCAGAACCTGCATATGGCGCAAAAAATATTTCAAACAACATTATCATATGTTGTTGAATGTTCAAATATTCAGTTACAACTCGAAGAAAACATGAAGAACATAAACGCTCTGAGAAAATATTTAAACATTATTTCAAAAAGTTAA
Protein
MGQSSVSTTKLPTESSPDFSASIMCKTIEEDGMTKLKLYQNKENTIVPLHWFGVLVPQNLHMAQKIFQTTLSYVVECSNIQLQLEENMKNINALRKYLNIISKS
Summary
Uniprot
H9JKU9
A0A0L7LR88
A0A2H1WJV8
A0A3S2NGH3
A0A2A4J7U8
A0A2W1BYN2
+ More
S4NK48 A0A1E1WGA4 A0A194PNB4 A0A194QZT2 A0A212FJJ6 A0A1L8DCR2 A0A1L8DC75 A0A1B0CFH3 A0A1B0D551 A0A023EFN7 A0A1S4F4U0 A0A023EHA3 A0A023EH14 A0A182G1E4 A0A1Q3FKF6 Q17FK3 A0A2P8Y5V9 U5ERZ7 D6WY99 J3JW75 A0A1Y1LFF2 B0XD73 A0A1W4UJD3 A0A023F6X4 A0A0M4EQJ7 A0A0V0GCW1 B4I423 A0A224XLB9 Q9VND4 Q4QPV1 B4QWX5 B4LZA7 B4PUT4 A0A1B0BBX9 B4KD82 A0A2R7WHM4 R4G422 A0A1A9UN93 A0A0P4VQA3 A0A1J1HUC5 A0A1A9YK68 B3MX59 B3P2F3 A0A1B0AAD0 W8B4H5 B4GF72 A0A3B0JVE7 A0A1B0G6V0 Q296G3 A0A162S3X4 A0A0P6FWF9 B4JZ26 A0A0P6H2W2 A0A0P4X9X1 B4GF74 B4NIP5 A0A1I8MT65 A0A1A9WJL2 A0A0L7QY74 A0A034VMD9 A0A0N0BET0 A0A2M4C1N5 A0A2M4AG70 A0A2M3Z5B2 A0A2M4C0E9 A0A2M3ZHR5 A0A1B6CZ24 A0A2M3Z575 A0A2M4C1K9 A0A1L8EBM3 A0A1L8ECX7 A0A182YQM2 T1DI60 A0A182F890 A0A2U4B2D6 F4W6P3 A0A0J7KBH5 H0VFK9 A0A182SFJ1 A0A250YBC3 A0A195ERI0
S4NK48 A0A1E1WGA4 A0A194PNB4 A0A194QZT2 A0A212FJJ6 A0A1L8DCR2 A0A1L8DC75 A0A1B0CFH3 A0A1B0D551 A0A023EFN7 A0A1S4F4U0 A0A023EHA3 A0A023EH14 A0A182G1E4 A0A1Q3FKF6 Q17FK3 A0A2P8Y5V9 U5ERZ7 D6WY99 J3JW75 A0A1Y1LFF2 B0XD73 A0A1W4UJD3 A0A023F6X4 A0A0M4EQJ7 A0A0V0GCW1 B4I423 A0A224XLB9 Q9VND4 Q4QPV1 B4QWX5 B4LZA7 B4PUT4 A0A1B0BBX9 B4KD82 A0A2R7WHM4 R4G422 A0A1A9UN93 A0A0P4VQA3 A0A1J1HUC5 A0A1A9YK68 B3MX59 B3P2F3 A0A1B0AAD0 W8B4H5 B4GF72 A0A3B0JVE7 A0A1B0G6V0 Q296G3 A0A162S3X4 A0A0P6FWF9 B4JZ26 A0A0P6H2W2 A0A0P4X9X1 B4GF74 B4NIP5 A0A1I8MT65 A0A1A9WJL2 A0A0L7QY74 A0A034VMD9 A0A0N0BET0 A0A2M4C1N5 A0A2M4AG70 A0A2M3Z5B2 A0A2M4C0E9 A0A2M3ZHR5 A0A1B6CZ24 A0A2M3Z575 A0A2M4C1K9 A0A1L8EBM3 A0A1L8ECX7 A0A182YQM2 T1DI60 A0A182F890 A0A2U4B2D6 F4W6P3 A0A0J7KBH5 H0VFK9 A0A182SFJ1 A0A250YBC3 A0A195ERI0
Pubmed
19121390
26227816
28756777
23622113
26354079
22118469
+ More
24945155 26483478 17510324 29403074 18362917 19820115 22516182 23537049 28004739 25474469 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 24495485 15632085 25315136 25348373 25244985 21719571 21993624 28087693
24945155 26483478 17510324 29403074 18362917 19820115 22516182 23537049 28004739 25474469 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27129103 24495485 15632085 25315136 25348373 25244985 21719571 21993624 28087693
EMBL
BABH01011526
JTDY01000264
KOB78013.1
ODYU01008814
SOQ52744.1
RSAL01000049
+ More
RVE50433.1 NWSH01002496 PCG68164.1 PCG68165.1 KZ149942 PZC76883.1 GAIX01013439 JAA79121.1 GDQN01005031 GDQN01003742 JAT86023.1 JAT87312.1 KQ459600 KPI94229.1 KQ460930 KPJ10794.1 AGBW02008256 OWR53906.1 GFDF01009838 JAV04246.1 GFDF01010011 JAV04073.1 AJWK01010106 AJVK01025073 GAPW01005365 JAC08233.1 JXUM01148643 GAPW01005364 KQ570607 JAC08234.1 KXJ68333.1 GAPW01005462 JAC08136.1 JXUM01136598 KQ568453 KXJ68993.1 GFDL01007053 JAV27992.1 CH477270 EAT45340.1 PYGN01000894 PSN39621.1 GANO01003486 JAB56385.1 KQ971357 EFA07742.1 APGK01041456 BT127493 KB740994 KB632280 AEE62455.1 ENN75931.1 ERL91303.1 GEZM01062413 GEZM01062412 GEZM01062411 JAV69737.1 DS232744 EDS45327.1 GBBI01001576 JAC17136.1 CP012526 ALC46622.1 GECL01000919 JAP05205.1 CH480821 EDW54966.1 GFTR01003169 JAW13257.1 AE014297 KX532074 AAF52009.1 ANY27884.1 BT023665 AAY85065.1 CM000364 EDX11737.1 CH940650 EDW68142.2 CM000160 EDW97741.1 JXJN01011744 CH933806 EDW14864.2 KK854838 PTY19142.1 ACPB03001249 GAHY01001720 JAA75790.1 GDKW01002576 JAI54019.1 CVRI01000014 CRK89753.1 CH902629 EDV35282.1 CH954181 EDV47903.1 GAMC01018359 JAB88196.1 CH479182 EDW34257.1 OUUW01000008 SPP84382.1 CCAG010013874 CM000070 EAL28495.2 LRGB01000084 KZS20993.1 GDIQ01041838 JAN52899.1 CH916378 EDV98641.1 GDIQ01025272 JAN69465.1 GDIP01244317 JAI79084.1 EDW34259.1 CH964272 EDW83759.1 KQ414697 KOC63501.1 GAKP01015692 GAKP01015690 GAKP01015688 GAKP01015686 GAKP01015684 GAKP01015682 JAC43262.1 KQ435824 KOX72072.1 GGFJ01010082 MBW59223.1 GGFK01006460 MBW39781.1 GGFM01002958 MBW23709.1 GGFJ01009648 MBW58789.1 GGFM01007356 MBW28107.1 GEDC01019212 GEDC01018693 GEDC01016358 JAS18086.1 JAS18605.1 JAS20940.1 GGFM01002930 MBW23681.1 GGFJ01010051 MBW59192.1 GFDG01002790 JAV16009.1 GFDG01002218 JAV16581.1 GAMD01002041 JAA99549.1 GL887707 EGI70119.1 LBMM01010208 KMQ87609.1 AAKN02007001 GFFW01003922 JAV40866.1 KQ982021 KYN30484.1
RVE50433.1 NWSH01002496 PCG68164.1 PCG68165.1 KZ149942 PZC76883.1 GAIX01013439 JAA79121.1 GDQN01005031 GDQN01003742 JAT86023.1 JAT87312.1 KQ459600 KPI94229.1 KQ460930 KPJ10794.1 AGBW02008256 OWR53906.1 GFDF01009838 JAV04246.1 GFDF01010011 JAV04073.1 AJWK01010106 AJVK01025073 GAPW01005365 JAC08233.1 JXUM01148643 GAPW01005364 KQ570607 JAC08234.1 KXJ68333.1 GAPW01005462 JAC08136.1 JXUM01136598 KQ568453 KXJ68993.1 GFDL01007053 JAV27992.1 CH477270 EAT45340.1 PYGN01000894 PSN39621.1 GANO01003486 JAB56385.1 KQ971357 EFA07742.1 APGK01041456 BT127493 KB740994 KB632280 AEE62455.1 ENN75931.1 ERL91303.1 GEZM01062413 GEZM01062412 GEZM01062411 JAV69737.1 DS232744 EDS45327.1 GBBI01001576 JAC17136.1 CP012526 ALC46622.1 GECL01000919 JAP05205.1 CH480821 EDW54966.1 GFTR01003169 JAW13257.1 AE014297 KX532074 AAF52009.1 ANY27884.1 BT023665 AAY85065.1 CM000364 EDX11737.1 CH940650 EDW68142.2 CM000160 EDW97741.1 JXJN01011744 CH933806 EDW14864.2 KK854838 PTY19142.1 ACPB03001249 GAHY01001720 JAA75790.1 GDKW01002576 JAI54019.1 CVRI01000014 CRK89753.1 CH902629 EDV35282.1 CH954181 EDV47903.1 GAMC01018359 JAB88196.1 CH479182 EDW34257.1 OUUW01000008 SPP84382.1 CCAG010013874 CM000070 EAL28495.2 LRGB01000084 KZS20993.1 GDIQ01041838 JAN52899.1 CH916378 EDV98641.1 GDIQ01025272 JAN69465.1 GDIP01244317 JAI79084.1 EDW34259.1 CH964272 EDW83759.1 KQ414697 KOC63501.1 GAKP01015692 GAKP01015690 GAKP01015688 GAKP01015686 GAKP01015684 GAKP01015682 JAC43262.1 KQ435824 KOX72072.1 GGFJ01010082 MBW59223.1 GGFK01006460 MBW39781.1 GGFM01002958 MBW23709.1 GGFJ01009648 MBW58789.1 GGFM01007356 MBW28107.1 GEDC01019212 GEDC01018693 GEDC01016358 JAS18086.1 JAS18605.1 JAS20940.1 GGFM01002930 MBW23681.1 GGFJ01010051 MBW59192.1 GFDG01002790 JAV16009.1 GFDG01002218 JAV16581.1 GAMD01002041 JAA99549.1 GL887707 EGI70119.1 LBMM01010208 KMQ87609.1 AAKN02007001 GFFW01003922 JAV40866.1 KQ982021 KYN30484.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000007151 UP000092461 UP000092462 UP000069940 UP000249989 UP000008820 UP000245037 UP000007266 UP000019118 UP000030742 UP000002320 UP000192221 UP000092553 UP000001292 UP000000803 UP000000304 UP000008792 UP000002282 UP000092460 UP000009192 UP000015103 UP000078200 UP000183832 UP000092443 UP000007801 UP000008711 UP000092445 UP000008744 UP000268350 UP000092444 UP000001819 UP000076858 UP000001070 UP000007798 UP000095301 UP000091820 UP000053825 UP000053105 UP000076408 UP000069272 UP000245320 UP000007755 UP000036403 UP000005447 UP000075901 UP000078541
UP000007151 UP000092461 UP000092462 UP000069940 UP000249989 UP000008820 UP000245037 UP000007266 UP000019118 UP000030742 UP000002320 UP000192221 UP000092553 UP000001292 UP000000803 UP000000304 UP000008792 UP000002282 UP000092460 UP000009192 UP000015103 UP000078200 UP000183832 UP000092443 UP000007801 UP000008711 UP000092445 UP000008744 UP000268350 UP000092444 UP000001819 UP000076858 UP000001070 UP000007798 UP000095301 UP000091820 UP000053825 UP000053105 UP000076408 UP000069272 UP000245320 UP000007755 UP000036403 UP000005447 UP000075901 UP000078541
Pfam
PF01813 ATP-synt_D
ProteinModelPortal
H9JKU9
A0A0L7LR88
A0A2H1WJV8
A0A3S2NGH3
A0A2A4J7U8
A0A2W1BYN2
+ More
S4NK48 A0A1E1WGA4 A0A194PNB4 A0A194QZT2 A0A212FJJ6 A0A1L8DCR2 A0A1L8DC75 A0A1B0CFH3 A0A1B0D551 A0A023EFN7 A0A1S4F4U0 A0A023EHA3 A0A023EH14 A0A182G1E4 A0A1Q3FKF6 Q17FK3 A0A2P8Y5V9 U5ERZ7 D6WY99 J3JW75 A0A1Y1LFF2 B0XD73 A0A1W4UJD3 A0A023F6X4 A0A0M4EQJ7 A0A0V0GCW1 B4I423 A0A224XLB9 Q9VND4 Q4QPV1 B4QWX5 B4LZA7 B4PUT4 A0A1B0BBX9 B4KD82 A0A2R7WHM4 R4G422 A0A1A9UN93 A0A0P4VQA3 A0A1J1HUC5 A0A1A9YK68 B3MX59 B3P2F3 A0A1B0AAD0 W8B4H5 B4GF72 A0A3B0JVE7 A0A1B0G6V0 Q296G3 A0A162S3X4 A0A0P6FWF9 B4JZ26 A0A0P6H2W2 A0A0P4X9X1 B4GF74 B4NIP5 A0A1I8MT65 A0A1A9WJL2 A0A0L7QY74 A0A034VMD9 A0A0N0BET0 A0A2M4C1N5 A0A2M4AG70 A0A2M3Z5B2 A0A2M4C0E9 A0A2M3ZHR5 A0A1B6CZ24 A0A2M3Z575 A0A2M4C1K9 A0A1L8EBM3 A0A1L8ECX7 A0A182YQM2 T1DI60 A0A182F890 A0A2U4B2D6 F4W6P3 A0A0J7KBH5 H0VFK9 A0A182SFJ1 A0A250YBC3 A0A195ERI0
S4NK48 A0A1E1WGA4 A0A194PNB4 A0A194QZT2 A0A212FJJ6 A0A1L8DCR2 A0A1L8DC75 A0A1B0CFH3 A0A1B0D551 A0A023EFN7 A0A1S4F4U0 A0A023EHA3 A0A023EH14 A0A182G1E4 A0A1Q3FKF6 Q17FK3 A0A2P8Y5V9 U5ERZ7 D6WY99 J3JW75 A0A1Y1LFF2 B0XD73 A0A1W4UJD3 A0A023F6X4 A0A0M4EQJ7 A0A0V0GCW1 B4I423 A0A224XLB9 Q9VND4 Q4QPV1 B4QWX5 B4LZA7 B4PUT4 A0A1B0BBX9 B4KD82 A0A2R7WHM4 R4G422 A0A1A9UN93 A0A0P4VQA3 A0A1J1HUC5 A0A1A9YK68 B3MX59 B3P2F3 A0A1B0AAD0 W8B4H5 B4GF72 A0A3B0JVE7 A0A1B0G6V0 Q296G3 A0A162S3X4 A0A0P6FWF9 B4JZ26 A0A0P6H2W2 A0A0P4X9X1 B4GF74 B4NIP5 A0A1I8MT65 A0A1A9WJL2 A0A0L7QY74 A0A034VMD9 A0A0N0BET0 A0A2M4C1N5 A0A2M4AG70 A0A2M3Z5B2 A0A2M4C0E9 A0A2M3ZHR5 A0A1B6CZ24 A0A2M3Z575 A0A2M4C1K9 A0A1L8EBM3 A0A1L8ECX7 A0A182YQM2 T1DI60 A0A182F890 A0A2U4B2D6 F4W6P3 A0A0J7KBH5 H0VFK9 A0A182SFJ1 A0A250YBC3 A0A195ERI0
Ontologies
GO
PANTHER
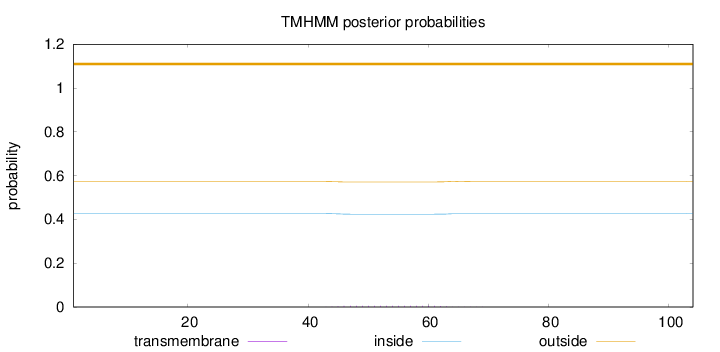
Topology
Length:
104
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10057
Exp number, first 60 AAs:
0.08425
Total prob of N-in:
0.42728
outside
1 - 104
Population Genetic Test Statistics
Pi
74.524966
Theta
122.926173
Tajima's D
-1.23632
CLR
0
CSRT
0.095745212739363
Interpretation
Uncertain
