Gene
KWMTBOMO03912
Pre Gene Modal
BGIBMGA010220
Annotation
hypothetical_protein_KGM_21654_[Danaus_plexippus]
Full name
Inhibitor of growth protein
Location in the cell
Nuclear Reliability : 4.39
Sequence
CDS
ATGTTAAACCAAACTTCGACTGAAGCTTTGCTTTCAGCCGCTTATGTTGAAAACTACTTAGATTGCGTAGAAAACCTTCCAAATGATCTTCAGAGACATTTATCACGTATGCGGGAGCTTGACGTAACATATAGGGAATGTTTGCGTGAGGCAGAAAAGCATCTAAATATTTGTATAGGATCAAGCACAGAGGAACGCCGTCGAACCAGAGCTGCACTACGCCTACAAACAGTTTTAGTTGCTGCACAAGAGATAGGAGATGAGAAACTACAAGTTGTACAAATATTACAAGATCTCATCGACAACAAACAAAGGTCACTTGATTCTGATCACAAAAAATTATTATCTTGTATAGAAGTAACAACAAATGGCACTACAAAAGAAGAGGTGACTCCAACTCAAGATGTACTCTCCAAGACAGAGAAGGAAGCCAGTGTGTTAACAACTCCTCATCAAAACATAGCACCACCGCCTCCTCCAGAAAGAACAAATGAGAAAGAAAAAGAAAAAGTTGAGAAGGAAAAAGGCAGTGGTGAACGGTGGTCAAAAAGACCACGTAGATCAAGAACAACAGCAGGTGCAATGACAGATGGAGCAGATTCCAGTGAACGAGATAGTGAGAGGCACACACACAACACAACACATAAGAAAGGTATAGGTAAAAAGAAAAAACGCAAAGTCCGACAAACGACGCAACGTTCGGAGACGCCACCGGAAGAGCCAGATGCCATCGACCCCGATGAGCCTCGCTACTGTCTCTGTGACCAAATCTCATTTGGCGAGATGATTCTCTGCGACAACGACTTATGTCCTATAGAGTGGTTCCACTTCTCGTGCGTTTCTCTCACAACTAAACCTAAAGGGAAATGGTTTTGTCCCAAGTGTCGCGGCGACAGACCGAATGTCATGAAGCCGAAGGGACAATTTCTTAAAGAATTAGAAAGATACAACAGAGAGAAAGAAGAAAAAGCATAG
Protein
MLNQTSTEALLSAAYVENYLDCVENLPNDLQRHLSRMRELDVTYRECLREAEKHLNICIGSSTEERRRTRAALRLQTVLVAAQEIGDEKLQVVQILQDLIDNKQRSLDSDHKKLLSCIEVTTNGTTKEEVTPTQDVLSKTEKEASVLTTPHQNIAPPPPPERTNEKEKEKVEKEKGSGERWSKRPRRSRTTAGAMTDGADSSERDSERHTHNTTHKKGIGKKKKRKVRQTTQRSETPPEEPDAIDPDEPRYCLCDQISFGEMILCDNDLCPIEWFHFSCVSLTTKPKGKWFCPKCRGDRPNVMKPKGQFLKELERYNREKEEKA
Summary
Subunit
Interacts with H3K4me3 and to a lesser extent with H3K4me2.
Similarity
Belongs to the ING family.
Feature
chain Inhibitor of growth protein
Uniprot
H9JL19
A0A212EJK2
A0A2W1BPC9
A0A2H1WEQ7
A0A2A4JRY7
A0A194PMM3
+ More
A0A194R0N5 A0A3S2PJM1 A0A3L8DYE1 A0A026WDL6 E2A2Q1 A0A0J7K3C3 A0A0L7RH20 A0A088ADL2 A0A232FNG7 E2C212 A0A2A3ENK0 A0A0K8TTN9 A0A0P5A3X2 A0A0N8C639 A0A0A9VVI3 A0A146KMZ2 A0A0P5ZHR7 A0A0P5ME96 A0A0N8BPR6 A0A0P5UQI4 A0A0N8EKG0 A0A1L8DWY5 A0A1B6FQA1 D2A1S8 A0A1B6CTU9 A0A0N8A268 A0A1Y1NFN4 A0A0P6GEK7 A0A0P5BXA8 A0A0P6GKG3 A0A0P5LA78 A0A0P5KYC2 A0A0P4Y765 A0A0P4Y1K7 A0A1W4X556 A0A0P5HQ18 C1BNW8 A0A0P6CIA7 A0A0P6EN00 A0A0P6FD74 A0A2S2QJH3 A0A0P4W150 J9K793 A0A2P6KBC6 A0A0N8C7I9 A0A0P5RE91 A0A0P5IJV3 A0A0P5IJE3 A0A0P5IPF2 A0A0P6CQ51 A0A0P5Y5G6
A0A194R0N5 A0A3S2PJM1 A0A3L8DYE1 A0A026WDL6 E2A2Q1 A0A0J7K3C3 A0A0L7RH20 A0A088ADL2 A0A232FNG7 E2C212 A0A2A3ENK0 A0A0K8TTN9 A0A0P5A3X2 A0A0N8C639 A0A0A9VVI3 A0A146KMZ2 A0A0P5ZHR7 A0A0P5ME96 A0A0N8BPR6 A0A0P5UQI4 A0A0N8EKG0 A0A1L8DWY5 A0A1B6FQA1 D2A1S8 A0A1B6CTU9 A0A0N8A268 A0A1Y1NFN4 A0A0P6GEK7 A0A0P5BXA8 A0A0P6GKG3 A0A0P5LA78 A0A0P5KYC2 A0A0P4Y765 A0A0P4Y1K7 A0A1W4X556 A0A0P5HQ18 C1BNW8 A0A0P6CIA7 A0A0P6EN00 A0A0P6FD74 A0A2S2QJH3 A0A0P4W150 J9K793 A0A2P6KBC6 A0A0N8C7I9 A0A0P5RE91 A0A0P5IJV3 A0A0P5IJE3 A0A0P5IPF2 A0A0P6CQ51 A0A0P5Y5G6
Pubmed
EMBL
BABH01011496
AGBW02014441
OWR41669.1
KZ149942
PZC76899.1
ODYU01008168
+ More
SOQ51549.1 NWSH01000744 PCG74458.1 KQ459600 KPI94243.1 KQ460930 KPJ10810.1 RSAL01000013 RVE53352.1 QOIP01000003 RLU24969.1 KK107295 EZA53104.1 GL436195 EFN72289.1 LBMM01015774 KMQ84671.1 KQ414596 KOC69996.1 NNAY01000009 OXU32069.1 GL452056 EFN78019.1 KZ288206 PBC33084.1 GDAI01000293 JAI17310.1 GDIP01204342 JAJ19060.1 GDIQ01111147 JAL40579.1 GBHO01045381 GBRD01013783 JAF98222.1 JAG52043.1 GDHC01021957 GDHC01013307 JAP96671.1 JAQ05322.1 GDIP01043514 LRGB01000024 JAM60201.1 KZS21458.1 GDIQ01156932 JAK94793.1 GDIQ01156931 JAK94794.1 GDIP01111650 JAL92064.1 GDIQ01017988 JAN76749.1 GFDF01003114 JAV10970.1 GECZ01017393 JAS52376.1 KQ971338 EFA02094.1 GEDC01030727 GEDC01026797 GEDC01020329 GEDC01014198 GEDC01008558 JAS06571.1 JAS10501.1 JAS16969.1 JAS23100.1 JAS28740.1 GDIP01189793 JAJ33609.1 GEZM01006541 JAV95730.1 GDIQ01044730 JAN50007.1 GDIP01193998 JAJ29404.1 GDIQ01042481 JAN52256.1 GDIQ01177469 JAK74256.1 GDIQ01177468 JAK74257.1 GDIP01233838 JAI89563.1 GDIP01233837 JAI89564.1 GDIQ01230009 JAK21716.1 BT076297 ACO10721.1 GDIQ01090807 JAN03930.1 GDIQ01061618 JAN33119.1 GDIQ01061617 JAN33120.1 GGMS01008702 MBY77905.1 GDRN01148127 JAI56636.1 ABLF02009770 MWRG01017196 PRD23628.1 GDIQ01107147 JAL44579.1 GDIQ01107148 JAL44578.1 GDIQ01214759 JAK36966.1 GDIQ01212657 JAK39068.1 GDIQ01212656 JAK39069.1 GDIP01010998 JAM92717.1 GDIP01062661 JAM41054.1
SOQ51549.1 NWSH01000744 PCG74458.1 KQ459600 KPI94243.1 KQ460930 KPJ10810.1 RSAL01000013 RVE53352.1 QOIP01000003 RLU24969.1 KK107295 EZA53104.1 GL436195 EFN72289.1 LBMM01015774 KMQ84671.1 KQ414596 KOC69996.1 NNAY01000009 OXU32069.1 GL452056 EFN78019.1 KZ288206 PBC33084.1 GDAI01000293 JAI17310.1 GDIP01204342 JAJ19060.1 GDIQ01111147 JAL40579.1 GBHO01045381 GBRD01013783 JAF98222.1 JAG52043.1 GDHC01021957 GDHC01013307 JAP96671.1 JAQ05322.1 GDIP01043514 LRGB01000024 JAM60201.1 KZS21458.1 GDIQ01156932 JAK94793.1 GDIQ01156931 JAK94794.1 GDIP01111650 JAL92064.1 GDIQ01017988 JAN76749.1 GFDF01003114 JAV10970.1 GECZ01017393 JAS52376.1 KQ971338 EFA02094.1 GEDC01030727 GEDC01026797 GEDC01020329 GEDC01014198 GEDC01008558 JAS06571.1 JAS10501.1 JAS16969.1 JAS23100.1 JAS28740.1 GDIP01189793 JAJ33609.1 GEZM01006541 JAV95730.1 GDIQ01044730 JAN50007.1 GDIP01193998 JAJ29404.1 GDIQ01042481 JAN52256.1 GDIQ01177469 JAK74256.1 GDIQ01177468 JAK74257.1 GDIP01233838 JAI89563.1 GDIP01233837 JAI89564.1 GDIQ01230009 JAK21716.1 BT076297 ACO10721.1 GDIQ01090807 JAN03930.1 GDIQ01061618 JAN33119.1 GDIQ01061617 JAN33120.1 GGMS01008702 MBY77905.1 GDRN01148127 JAI56636.1 ABLF02009770 MWRG01017196 PRD23628.1 GDIQ01107147 JAL44579.1 GDIQ01107148 JAL44578.1 GDIQ01214759 JAK36966.1 GDIQ01212657 JAK39068.1 GDIQ01212656 JAK39069.1 GDIP01010998 JAM92717.1 GDIP01062661 JAM41054.1
Proteomes
PRIDE
Pfam
PF12998 ING
Interpro
SUPFAM
SSF57903
SSF57903
Gene 3D
ProteinModelPortal
H9JL19
A0A212EJK2
A0A2W1BPC9
A0A2H1WEQ7
A0A2A4JRY7
A0A194PMM3
+ More
A0A194R0N5 A0A3S2PJM1 A0A3L8DYE1 A0A026WDL6 E2A2Q1 A0A0J7K3C3 A0A0L7RH20 A0A088ADL2 A0A232FNG7 E2C212 A0A2A3ENK0 A0A0K8TTN9 A0A0P5A3X2 A0A0N8C639 A0A0A9VVI3 A0A146KMZ2 A0A0P5ZHR7 A0A0P5ME96 A0A0N8BPR6 A0A0P5UQI4 A0A0N8EKG0 A0A1L8DWY5 A0A1B6FQA1 D2A1S8 A0A1B6CTU9 A0A0N8A268 A0A1Y1NFN4 A0A0P6GEK7 A0A0P5BXA8 A0A0P6GKG3 A0A0P5LA78 A0A0P5KYC2 A0A0P4Y765 A0A0P4Y1K7 A0A1W4X556 A0A0P5HQ18 C1BNW8 A0A0P6CIA7 A0A0P6EN00 A0A0P6FD74 A0A2S2QJH3 A0A0P4W150 J9K793 A0A2P6KBC6 A0A0N8C7I9 A0A0P5RE91 A0A0P5IJV3 A0A0P5IJE3 A0A0P5IPF2 A0A0P6CQ51 A0A0P5Y5G6
A0A194R0N5 A0A3S2PJM1 A0A3L8DYE1 A0A026WDL6 E2A2Q1 A0A0J7K3C3 A0A0L7RH20 A0A088ADL2 A0A232FNG7 E2C212 A0A2A3ENK0 A0A0K8TTN9 A0A0P5A3X2 A0A0N8C639 A0A0A9VVI3 A0A146KMZ2 A0A0P5ZHR7 A0A0P5ME96 A0A0N8BPR6 A0A0P5UQI4 A0A0N8EKG0 A0A1L8DWY5 A0A1B6FQA1 D2A1S8 A0A1B6CTU9 A0A0N8A268 A0A1Y1NFN4 A0A0P6GEK7 A0A0P5BXA8 A0A0P6GKG3 A0A0P5LA78 A0A0P5KYC2 A0A0P4Y765 A0A0P4Y1K7 A0A1W4X556 A0A0P5HQ18 C1BNW8 A0A0P6CIA7 A0A0P6EN00 A0A0P6FD74 A0A2S2QJH3 A0A0P4W150 J9K793 A0A2P6KBC6 A0A0N8C7I9 A0A0P5RE91 A0A0P5IJV3 A0A0P5IJE3 A0A0P5IPF2 A0A0P6CQ51 A0A0P5Y5G6
PDB
1WES
E-value=7.97931e-22,
Score=255
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
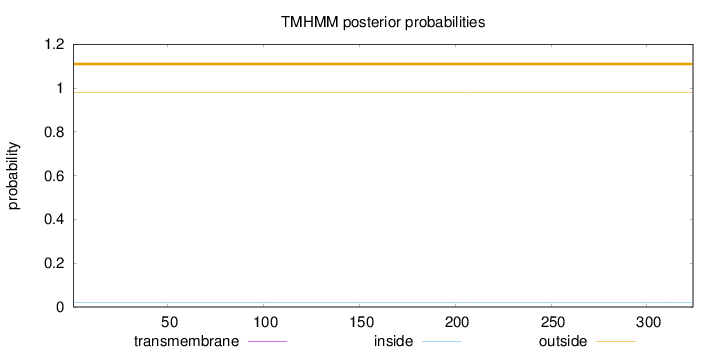
Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02084
outside
1 - 324
Population Genetic Test Statistics
Pi
307.286508
Theta
210.107458
Tajima's D
1.381553
CLR
0.143218
CSRT
0.765611719414029
Interpretation
Uncertain
