Pre Gene Modal
BGIBMGA010158
Annotation
PREDICTED:_microtubule-associated_protein_Jupiter_isoform_X2_[Papilio_xuthus]
Full name
Microtubule-associated protein Jupiter
Location in the cell
Nuclear Reliability : 4.095
Sequence
CDS
ATGACAGACGGCAGCCCCAAGAAGAACGGCATGGCGCGCCAACGTGTCGTCCGCGACACACCGACGCGTCCGCGTGACACTCACTCGAGACTTTTCGGACAGGCAGCGAGCAATGGACCCGTTTCTCCATTGATCACCGACACCATCCGCAGTAACATCCAGTTTGGTGATTCTGAAATGAACGGCGGAAGTCCTACGCACTCCCCCGCCAAGATGATGAACGGAAGCGCTTATTCGACACCCAGCCGAGCGTCATACACGCCACCGAAGAGGAATCCAGTGACAGGCGATGGAGTGCCTCTGACGCCGCTGCGTCGCCGTCATCCGGCTGCGAGCCTGCGAGAAGGCAACCCAATCACCGGTGACGGATACAAGTCGATGAACGGGCAAATGAACACAATGACGTCAATTAATGGCACCAGCAACATTATCTACTACAACCGCGTGCCGCCCGGCGGTTATTCATCTGGACTTTGGTAG
Protein
MTDGSPKKNGMARQRVVRDTPTRPRDTHSRLFGQAASNGPVSPLITDTIRSNIQFGDSEMNGGSPTHSPAKMMNGSAYSTPSRASYTPPKRNPVTGDGVPLTPLRRRHPAASLREGNPITGDGYKSMNGQMNTMTSINGTSNIIYYNRVPPGGYSSGLW
Summary
Description
Binds to all microtubule populations.
Similarity
Belongs to the MAP Jupiter family.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Nucleus
Phosphoprotein
Reference proteome
Feature
chain Microtubule-associated protein Jupiter
Uniprot
A0A3S2NKN0
H9JKV7
A0A2A4JMX6
A0A2A4JP24
I4DJD0
A0A2H1WFN2
+ More
A0A2W1BVS5 A0A194R4M4 A0A194PNC8 A0A212FDQ8 A0A1L8E5T8 A0A1L8E5C3 A0A067RAR3 A0A1L8E610 A0A1L8E5U6 A0A084VT96 A0A182N4A2 A0A0Q9X6Y6 Q4VIW8 Q4VIX0 B4KBL1 A0A0Q9WWW1 A0A0Q9WWW2 Q4VIW7 Q4VIW9 A0A0Q9WX33 A0A0Q9WVB5 A0A0Q9WTK8 A0A0Q9WXH7 A0A1I8QDL7 A0A0K8TM79 A0A1Q3FUY6 A0A1Q3FUW2 A0A0Q9X5A1 A0A182P2J0 W8AJM7 B4JUG8 A0A1Q3FUQ7 A0A1I8QDM6 A0A1W4W1C1 A0A0Q9WT98 A0A1Q3FUK9 A0A1W4WC14 A0A182F927 A0A034VTK9 A0A0K8W0H1 A0A034VXQ8 U5ERZ6 A0A2M4CJ18 A0A0K8V1I7
A0A2W1BVS5 A0A194R4M4 A0A194PNC8 A0A212FDQ8 A0A1L8E5T8 A0A1L8E5C3 A0A067RAR3 A0A1L8E610 A0A1L8E5U6 A0A084VT96 A0A182N4A2 A0A0Q9X6Y6 Q4VIW8 Q4VIX0 B4KBL1 A0A0Q9WWW1 A0A0Q9WWW2 Q4VIW7 Q4VIW9 A0A0Q9WX33 A0A0Q9WVB5 A0A0Q9WTK8 A0A0Q9WXH7 A0A1I8QDL7 A0A0K8TM79 A0A1Q3FUY6 A0A1Q3FUW2 A0A0Q9X5A1 A0A182P2J0 W8AJM7 B4JUG8 A0A1Q3FUQ7 A0A1I8QDM6 A0A1W4W1C1 A0A0Q9WT98 A0A1Q3FUK9 A0A1W4WC14 A0A182F927 A0A034VTK9 A0A0K8W0H1 A0A034VXQ8 U5ERZ6 A0A2M4CJ18 A0A0K8V1I7
Pubmed
EMBL
RSAL01000013
RVE53353.1
BABH01011486
NWSH01000998
PCG73169.1
PCG73172.1
+ More
AK401398 BAM18020.1 ODYU01008367 SOQ51895.1 KZ149942 PZC76900.1 KQ460930 KPJ10811.1 KQ459600 KPI94244.1 AGBW02009013 OWR51864.1 GFDF01000011 JAV14073.1 GFDF01000265 JAV13819.1 KK852818 KDR15720.1 GFDF01000005 JAV14079.1 GFDF01000006 JAV14078.1 ATLV01016283 KE525074 KFB41190.1 CH933806 KRG00558.1 AY900631 AAX12429.1 AAX12428.1 EDW13678.1 KRG00559.1 KRG00560.1 AAX12430.1 AAX12427.1 KRG00556.1 CH964232 KRF99448.1 KRF99451.1 KRG00557.1 GDAI01002568 JAI15035.1 GFDL01003792 JAV31253.1 GFDL01003769 JAV31276.1 KRF99447.1 GAMC01021642 JAB84913.1 CH916374 EDV91138.1 GFDL01003760 JAV31285.1 KRF99450.1 GFDL01003756 JAV31289.1 GAKP01012306 JAC46646.1 GDHF01007989 JAI44325.1 GAKP01012302 JAC46650.1 GANO01002597 JAB57274.1 GGFL01001172 MBW65350.1 GDHF01019656 JAI32658.1
AK401398 BAM18020.1 ODYU01008367 SOQ51895.1 KZ149942 PZC76900.1 KQ460930 KPJ10811.1 KQ459600 KPI94244.1 AGBW02009013 OWR51864.1 GFDF01000011 JAV14073.1 GFDF01000265 JAV13819.1 KK852818 KDR15720.1 GFDF01000005 JAV14079.1 GFDF01000006 JAV14078.1 ATLV01016283 KE525074 KFB41190.1 CH933806 KRG00558.1 AY900631 AAX12429.1 AAX12428.1 EDW13678.1 KRG00559.1 KRG00560.1 AAX12430.1 AAX12427.1 KRG00556.1 CH964232 KRF99448.1 KRF99451.1 KRG00557.1 GDAI01002568 JAI15035.1 GFDL01003792 JAV31253.1 GFDL01003769 JAV31276.1 KRF99447.1 GAMC01021642 JAB84913.1 CH916374 EDV91138.1 GFDL01003760 JAV31285.1 KRF99450.1 GFDL01003756 JAV31289.1 GAKP01012306 JAC46646.1 GDHF01007989 JAI44325.1 GAKP01012302 JAC46650.1 GANO01002597 JAB57274.1 GGFL01001172 MBW65350.1 GDHF01019656 JAI32658.1
Proteomes
Pfam
PF17054 JUPITER
Interpro
IPR033335
JUPITER
ProteinModelPortal
A0A3S2NKN0
H9JKV7
A0A2A4JMX6
A0A2A4JP24
I4DJD0
A0A2H1WFN2
+ More
A0A2W1BVS5 A0A194R4M4 A0A194PNC8 A0A212FDQ8 A0A1L8E5T8 A0A1L8E5C3 A0A067RAR3 A0A1L8E610 A0A1L8E5U6 A0A084VT96 A0A182N4A2 A0A0Q9X6Y6 Q4VIW8 Q4VIX0 B4KBL1 A0A0Q9WWW1 A0A0Q9WWW2 Q4VIW7 Q4VIW9 A0A0Q9WX33 A0A0Q9WVB5 A0A0Q9WTK8 A0A0Q9WXH7 A0A1I8QDL7 A0A0K8TM79 A0A1Q3FUY6 A0A1Q3FUW2 A0A0Q9X5A1 A0A182P2J0 W8AJM7 B4JUG8 A0A1Q3FUQ7 A0A1I8QDM6 A0A1W4W1C1 A0A0Q9WT98 A0A1Q3FUK9 A0A1W4WC14 A0A182F927 A0A034VTK9 A0A0K8W0H1 A0A034VXQ8 U5ERZ6 A0A2M4CJ18 A0A0K8V1I7
A0A2W1BVS5 A0A194R4M4 A0A194PNC8 A0A212FDQ8 A0A1L8E5T8 A0A1L8E5C3 A0A067RAR3 A0A1L8E610 A0A1L8E5U6 A0A084VT96 A0A182N4A2 A0A0Q9X6Y6 Q4VIW8 Q4VIX0 B4KBL1 A0A0Q9WWW1 A0A0Q9WWW2 Q4VIW7 Q4VIW9 A0A0Q9WX33 A0A0Q9WVB5 A0A0Q9WTK8 A0A0Q9WXH7 A0A1I8QDL7 A0A0K8TM79 A0A1Q3FUY6 A0A1Q3FUW2 A0A0Q9X5A1 A0A182P2J0 W8AJM7 B4JUG8 A0A1Q3FUQ7 A0A1I8QDM6 A0A1W4W1C1 A0A0Q9WT98 A0A1Q3FUK9 A0A1W4WC14 A0A182F927 A0A034VTK9 A0A0K8W0H1 A0A034VXQ8 U5ERZ6 A0A2M4CJ18 A0A0K8V1I7
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoskeleton
Spindle
Cytoplasm
Cytoskeleton
Spindle
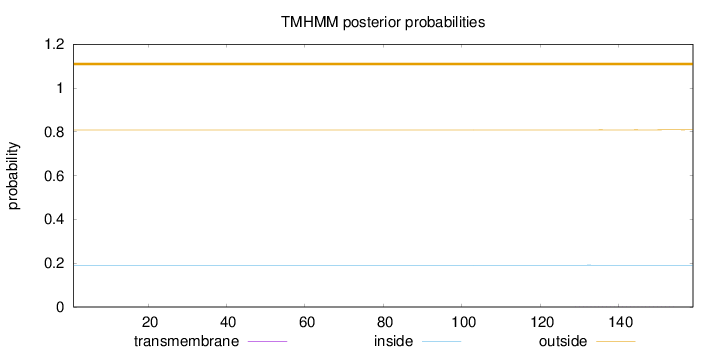
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01198
Exp number, first 60 AAs:
0.00071
Total prob of N-in:
0.19124
outside
1 - 159
Population Genetic Test Statistics
Pi
269.532208
Theta
186.141873
Tajima's D
1.398307
CLR
0.249233
CSRT
0.76341182940853
Interpretation
Uncertain
