Gene
KWMTBOMO03904
Pre Gene Modal
BGIBMGA010162
Annotation
Facilitated_trehalose_transporter_Tret1_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.98
Sequence
CDS
ATGTCTCCAAACTTACTTCTGTTGGATCTTGGTATGGCCGTAAGCTTCCCAACCATAGCTTTACCGGCTTTGCTGAATGCCACAGAAGGATTGTCTCAGACGCAAGTAGAGGCTTCGTGGTTTGGTAGCATTTCATATCTTGCTCAACCTTTTGGGGCTCTGCTATCTGGTCCACTTGTAGACTACTTTGGAAGGAAAAAAGCAAACTTTTTAGTTAATATACCTCATTTAATAGCTTGGATCTTGATGTATTTTGCCTGGAATGTACCGATTTTATTTATAGCAAATACTCTGTTAGGGATTGGAACCGGGATAATGGAAGCGCCAATAAATTCTTATGTTGGGGAAATTAGTGAGCCCACGATGCGTGGAGCCCTATGCACCGTCACTCAAATATTCACGTCCATTGGAATATTGGTAATGTATTTTCTGGGAACAATAGTAGATTGGCGACAAGCGGCAATCATATGTCTAGGAGTACCACTTCTTTCCATGCTATTAGTTTTACTTGTACCCGAAACTCCTGTCTGGCTACTGTTCAAAGGGAGAGAAAAAGAAGCATTAAAATCTTTGTGCTACCTTCGAGGGTGGACAACTGTTGACAATGTAAAGGACGAATATGATAAATTAGCAGTTTATGCGAAGAGTTTGCAAAACTGCGAAATCTGTCTCGCCACTGATGACGTTAAAATTGAGCAATGTGTCCACGCTCGAATGAATCCAGTAAAAAGATACATCTTAAAGATCAAATGCGTAATGTTCGGCAAAGAGACTCTAAGGCCATTAACATTGGTAATTCTATATTTTATGTTCTACACTATGAGTGGACTCATACCTATTAGACCTAATATGGTCAATGTATGTGGAGCCTTCGGGATGGTGGACAATGGAAAGAAGATAGTGCTAATGATCGGTGTTATAACTGTAATCATGGGCGTCATAGTCATCGGTATCATCAAAATGACCGGTAAACGAAAATTGATTATACTAGCGCTTCTCGGCACTGGTCTTTCCAGCACTGGACTAAGTATATACGCAAGATTACACGTACCTGATTCGGTGTCTTCATACGATCCTGCCACATTTCCTGTAGAAAAAAGCTACATACCTCAGGCGCTATTGTATTCCTTGGCGGTGTTTACTGGGCTCTGTATACCTTGGGTGCTGCTTGGAGAAATGTTCCCTTTTAAGAGTCGTGCCTCAGCTCAAGGAATCGCTGCTGCTTGGAGTTACGTGGTGACGTTCTTGGGATCAAAAACGTTAATCGATTTGGAGAATTCGCTTAAGCTGTGGGGAACTTTTGCCGTTTATGCAGTTTTCGCATTTGTTGGCACAATATATTTATATTTCTTCATGCCTGAAACTGAAGGGAAGACGCTTCAAGACATCGAAGATTATTATAAAGGGGAATTGAGAATATTCGCGAACGATCCTTTTATAAATTACTTTAAGAGATTCAAGAGAAAACAGTCTGTCTCTTAG
Protein
MSPNLLLLDLGMAVSFPTIALPALLNATEGLSQTQVEASWFGSISYLAQPFGALLSGPLVDYFGRKKANFLVNIPHLIAWILMYFAWNVPILFIANTLLGIGTGIMEAPINSYVGEISEPTMRGALCTVTQIFTSIGILVMYFLGTIVDWRQAAIICLGVPLLSMLLVLLVPETPVWLLFKGREKEALKSLCYLRGWTTVDNVKDEYDKLAVYAKSLQNCEICLATDDVKIEQCVHARMNPVKRYILKIKCVMFGKETLRPLTLVILYFMFYTMSGLIPIRPNMVNVCGAFGMVDNGKKIVLMIGVITVIMGVIVIGIIKMTGKRKLIILALLGTGLSSTGLSIYARLHVPDSVSSYDPATFPVEKSYIPQALLYSLAVFTGLCIPWVLLGEMFPFKSRASAQGIAAAWSYVVTFLGSKTLIDLENSLKLWGTFAVYAVFAFVGTIYLYFFMPETEGKTLQDIEDYYKGELRIFANDPFINYFKRFKRKQSVS
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A0N1IPR7
A0A194PLE0
H9JKW0
A0A194PSZ3
A0A0N1PJU5
A0A2H1WJR8
+ More
A0A212F4D1 A0A2A4K0B8 A0A212FGM4 A0A0L7KVV7 A0A3S2LTC9 A0A212FGM3 A0A194PMM6 A0A2A4K146 A0A0L7KQ05 A0A2W1BPD0 A0A2W1B4Q0 A0A182FS13 A0A2M3Z0P0 A0A2M3ZG53 A0A1L8DF11 A0A2M3Z0I1 A0A1L8DFC7 A0A2M4AGT5 A0A2M4AH60 A0A2M4BIQ4 A0A2M4BIL1 A0A2M4BJE1 W5J8E4 B0WLM4 A0A182V5A4 Q7PX65 A0A182X9E6 A0A182U3Q5 A0A182HKD2 A0A1Q3FRP9 A0A182RSB7 A0A182IUY9 A0A182R125 A0A1B0CIY2 Q16TJ6 A0A182NQQ7 A0A182YHY6 A0A182VVZ1 A0A182JVE9 A0A182GM49 A0A023ETX2 A0A1B0CIY3 A0A182PRT8 A0A2J7PIK9 A0A182MTT7 A0A1L8DF49 A0A0K8TQS7 U5EU35 A0A2P8Y8K3 A0A1B0C8F7 A0A1B6HSC1 A0A182L8L4 A0A1B0CIY4 A0A1B6BX46 A0A1B6IZ61 A0A2P8ZNY9 A0A067RQY9 A0A0A8J7Z9 A0A0A8J8M1 A0A067QV41 A0A2J7PIN1 A0A1B6H9R4 A0A1B0CIY0 A0A2J7QL19 A0A2H8TCU3 A0A2J7QL26 A0A023EZ96 A0A1B6EU41 A0A1B6M6C7 A0A1B6JJ73 A0A2J7QC67 A0A1B6E8P1 A0A146LN52 A0A224XL30 A0A0A9ZCC5 A0A2J7QC75 T1IC40 A0A069DUA8 A0A2P8XHU0 T1HYD0 A0A2P8Y8I9 A0A224XK01 D4AHX2 A0A1B6M0V4 A0A1B6IEH6 A0A1S3D6K1 A0A1B6GQD0 A0A146KUE8
A0A212F4D1 A0A2A4K0B8 A0A212FGM4 A0A0L7KVV7 A0A3S2LTC9 A0A212FGM3 A0A194PMM6 A0A2A4K146 A0A0L7KQ05 A0A2W1BPD0 A0A2W1B4Q0 A0A182FS13 A0A2M3Z0P0 A0A2M3ZG53 A0A1L8DF11 A0A2M3Z0I1 A0A1L8DFC7 A0A2M4AGT5 A0A2M4AH60 A0A2M4BIQ4 A0A2M4BIL1 A0A2M4BJE1 W5J8E4 B0WLM4 A0A182V5A4 Q7PX65 A0A182X9E6 A0A182U3Q5 A0A182HKD2 A0A1Q3FRP9 A0A182RSB7 A0A182IUY9 A0A182R125 A0A1B0CIY2 Q16TJ6 A0A182NQQ7 A0A182YHY6 A0A182VVZ1 A0A182JVE9 A0A182GM49 A0A023ETX2 A0A1B0CIY3 A0A182PRT8 A0A2J7PIK9 A0A182MTT7 A0A1L8DF49 A0A0K8TQS7 U5EU35 A0A2P8Y8K3 A0A1B0C8F7 A0A1B6HSC1 A0A182L8L4 A0A1B0CIY4 A0A1B6BX46 A0A1B6IZ61 A0A2P8ZNY9 A0A067RQY9 A0A0A8J7Z9 A0A0A8J8M1 A0A067QV41 A0A2J7PIN1 A0A1B6H9R4 A0A1B0CIY0 A0A2J7QL19 A0A2H8TCU3 A0A2J7QL26 A0A023EZ96 A0A1B6EU41 A0A1B6M6C7 A0A1B6JJ73 A0A2J7QC67 A0A1B6E8P1 A0A146LN52 A0A224XL30 A0A0A9ZCC5 A0A2J7QC75 T1IC40 A0A069DUA8 A0A2P8XHU0 T1HYD0 A0A2P8Y8I9 A0A224XK01 D4AHX2 A0A1B6M0V4 A0A1B6IEH6 A0A1S3D6K1 A0A1B6GQD0 A0A146KUE8
Pubmed
EMBL
KQ460317
KPJ15996.1
KQ459600
KPI94251.1
BABH01011474
BABH01011475
+ More
KPI94250.1 KPJ15995.1 ODYU01009059 SOQ53206.1 AGBW02010404 OWR48573.1 NWSH01000329 PCG77368.1 AGBW02008643 OWR52884.1 JTDY01005248 KOB67201.1 RSAL01000013 RVE53405.1 OWR52886.1 KPI94248.1 PCG77362.1 JTDY01007184 KOB65398.1 KZ149942 PZC76902.1 KZ150329 PZC71362.1 GGFM01001257 MBW22008.1 GGFM01006677 MBW27428.1 GFDF01009031 JAV05053.1 GGFM01001274 MBW22025.1 GFDF01009024 JAV05060.1 GGFK01006601 MBW39922.1 GGFK01006805 MBW40126.1 GGFJ01003743 MBW52884.1 GGFJ01003736 MBW52877.1 GGFJ01003767 MBW52908.1 ADMH02002101 ETN59145.1 DS231988 EDS30566.1 AAAB01008987 EAA01784.5 APCN01000826 GFDL01004766 JAV30279.1 AXCN02001503 AJWK01013897 AJWK01013898 CH477648 EAT37826.1 JXUM01073814 JXUM01073815 KQ562796 KXJ75112.1 GAPW01000773 JAC12825.1 AJWK01013900 NEVH01025126 PNF16170.1 AXCM01002705 GFDF01009002 JAV05082.1 GDAI01000891 JAI16712.1 GANO01004060 JAB55811.1 PYGN01000805 PSN40582.1 AJWK01000542 GECU01030165 JAS77541.1 AJWK01013901 GEDC01031445 JAS05853.1 GECU01015504 JAS92202.1 PYGN01000006 PSN58216.1 KK852475 KDR23065.1 AB901072 BAQ02372.1 AB901059 BAQ02359.1 KK853229 KDR09649.1 PNF16169.1 GECU01036228 GECU01032505 JAS71478.1 JAS75201.1 AJWK01013892 AJWK01013893 NEVH01013254 PNF29284.1 GFXV01000110 MBW11915.1 PNF29286.1 GBBI01004167 JAC14545.1 GECZ01028516 JAS41253.1 GEBQ01008509 JAT31468.1 GECU01008471 JAS99235.1 NEVH01016291 PNF26177.1 GEDC01002997 JAS34301.1 GDHC01010522 JAQ08107.1 GFTR01007114 JAW09312.1 GBHO01000682 JAG42922.1 PNF26178.1 ACPB03000842 GBGD01001176 JAC87713.1 PYGN01002079 PSN31571.1 ACPB03023997 PSN40576.1 GFTR01006278 JAW10148.1 AB550000 BAI83421.1 GEBQ01010411 JAT29566.1 GECU01022368 JAS85338.1 GECZ01005138 JAS64631.1 GDHC01018701 JAP99927.1
KPI94250.1 KPJ15995.1 ODYU01009059 SOQ53206.1 AGBW02010404 OWR48573.1 NWSH01000329 PCG77368.1 AGBW02008643 OWR52884.1 JTDY01005248 KOB67201.1 RSAL01000013 RVE53405.1 OWR52886.1 KPI94248.1 PCG77362.1 JTDY01007184 KOB65398.1 KZ149942 PZC76902.1 KZ150329 PZC71362.1 GGFM01001257 MBW22008.1 GGFM01006677 MBW27428.1 GFDF01009031 JAV05053.1 GGFM01001274 MBW22025.1 GFDF01009024 JAV05060.1 GGFK01006601 MBW39922.1 GGFK01006805 MBW40126.1 GGFJ01003743 MBW52884.1 GGFJ01003736 MBW52877.1 GGFJ01003767 MBW52908.1 ADMH02002101 ETN59145.1 DS231988 EDS30566.1 AAAB01008987 EAA01784.5 APCN01000826 GFDL01004766 JAV30279.1 AXCN02001503 AJWK01013897 AJWK01013898 CH477648 EAT37826.1 JXUM01073814 JXUM01073815 KQ562796 KXJ75112.1 GAPW01000773 JAC12825.1 AJWK01013900 NEVH01025126 PNF16170.1 AXCM01002705 GFDF01009002 JAV05082.1 GDAI01000891 JAI16712.1 GANO01004060 JAB55811.1 PYGN01000805 PSN40582.1 AJWK01000542 GECU01030165 JAS77541.1 AJWK01013901 GEDC01031445 JAS05853.1 GECU01015504 JAS92202.1 PYGN01000006 PSN58216.1 KK852475 KDR23065.1 AB901072 BAQ02372.1 AB901059 BAQ02359.1 KK853229 KDR09649.1 PNF16169.1 GECU01036228 GECU01032505 JAS71478.1 JAS75201.1 AJWK01013892 AJWK01013893 NEVH01013254 PNF29284.1 GFXV01000110 MBW11915.1 PNF29286.1 GBBI01004167 JAC14545.1 GECZ01028516 JAS41253.1 GEBQ01008509 JAT31468.1 GECU01008471 JAS99235.1 NEVH01016291 PNF26177.1 GEDC01002997 JAS34301.1 GDHC01010522 JAQ08107.1 GFTR01007114 JAW09312.1 GBHO01000682 JAG42922.1 PNF26178.1 ACPB03000842 GBGD01001176 JAC87713.1 PYGN01002079 PSN31571.1 ACPB03023997 PSN40576.1 GFTR01006278 JAW10148.1 AB550000 BAI83421.1 GEBQ01010411 JAT29566.1 GECU01022368 JAS85338.1 GECZ01005138 JAS64631.1 GDHC01018701 JAP99927.1
Proteomes
UP000053240
UP000053268
UP000005204
UP000007151
UP000218220
UP000037510
+ More
UP000283053 UP000069272 UP000000673 UP000002320 UP000075903 UP000007062 UP000076407 UP000075902 UP000075840 UP000075900 UP000075880 UP000075886 UP000092461 UP000008820 UP000075884 UP000076408 UP000075920 UP000075881 UP000069940 UP000249989 UP000075885 UP000235965 UP000075883 UP000245037 UP000075882 UP000027135 UP000015103 UP000079169
UP000283053 UP000069272 UP000000673 UP000002320 UP000075903 UP000007062 UP000076407 UP000075902 UP000075840 UP000075900 UP000075880 UP000075886 UP000092461 UP000008820 UP000075884 UP000076408 UP000075920 UP000075881 UP000069940 UP000249989 UP000075885 UP000235965 UP000075883 UP000245037 UP000075882 UP000027135 UP000015103 UP000079169
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A0N1IPR7
A0A194PLE0
H9JKW0
A0A194PSZ3
A0A0N1PJU5
A0A2H1WJR8
+ More
A0A212F4D1 A0A2A4K0B8 A0A212FGM4 A0A0L7KVV7 A0A3S2LTC9 A0A212FGM3 A0A194PMM6 A0A2A4K146 A0A0L7KQ05 A0A2W1BPD0 A0A2W1B4Q0 A0A182FS13 A0A2M3Z0P0 A0A2M3ZG53 A0A1L8DF11 A0A2M3Z0I1 A0A1L8DFC7 A0A2M4AGT5 A0A2M4AH60 A0A2M4BIQ4 A0A2M4BIL1 A0A2M4BJE1 W5J8E4 B0WLM4 A0A182V5A4 Q7PX65 A0A182X9E6 A0A182U3Q5 A0A182HKD2 A0A1Q3FRP9 A0A182RSB7 A0A182IUY9 A0A182R125 A0A1B0CIY2 Q16TJ6 A0A182NQQ7 A0A182YHY6 A0A182VVZ1 A0A182JVE9 A0A182GM49 A0A023ETX2 A0A1B0CIY3 A0A182PRT8 A0A2J7PIK9 A0A182MTT7 A0A1L8DF49 A0A0K8TQS7 U5EU35 A0A2P8Y8K3 A0A1B0C8F7 A0A1B6HSC1 A0A182L8L4 A0A1B0CIY4 A0A1B6BX46 A0A1B6IZ61 A0A2P8ZNY9 A0A067RQY9 A0A0A8J7Z9 A0A0A8J8M1 A0A067QV41 A0A2J7PIN1 A0A1B6H9R4 A0A1B0CIY0 A0A2J7QL19 A0A2H8TCU3 A0A2J7QL26 A0A023EZ96 A0A1B6EU41 A0A1B6M6C7 A0A1B6JJ73 A0A2J7QC67 A0A1B6E8P1 A0A146LN52 A0A224XL30 A0A0A9ZCC5 A0A2J7QC75 T1IC40 A0A069DUA8 A0A2P8XHU0 T1HYD0 A0A2P8Y8I9 A0A224XK01 D4AHX2 A0A1B6M0V4 A0A1B6IEH6 A0A1S3D6K1 A0A1B6GQD0 A0A146KUE8
A0A212F4D1 A0A2A4K0B8 A0A212FGM4 A0A0L7KVV7 A0A3S2LTC9 A0A212FGM3 A0A194PMM6 A0A2A4K146 A0A0L7KQ05 A0A2W1BPD0 A0A2W1B4Q0 A0A182FS13 A0A2M3Z0P0 A0A2M3ZG53 A0A1L8DF11 A0A2M3Z0I1 A0A1L8DFC7 A0A2M4AGT5 A0A2M4AH60 A0A2M4BIQ4 A0A2M4BIL1 A0A2M4BJE1 W5J8E4 B0WLM4 A0A182V5A4 Q7PX65 A0A182X9E6 A0A182U3Q5 A0A182HKD2 A0A1Q3FRP9 A0A182RSB7 A0A182IUY9 A0A182R125 A0A1B0CIY2 Q16TJ6 A0A182NQQ7 A0A182YHY6 A0A182VVZ1 A0A182JVE9 A0A182GM49 A0A023ETX2 A0A1B0CIY3 A0A182PRT8 A0A2J7PIK9 A0A182MTT7 A0A1L8DF49 A0A0K8TQS7 U5EU35 A0A2P8Y8K3 A0A1B0C8F7 A0A1B6HSC1 A0A182L8L4 A0A1B0CIY4 A0A1B6BX46 A0A1B6IZ61 A0A2P8ZNY9 A0A067RQY9 A0A0A8J7Z9 A0A0A8J8M1 A0A067QV41 A0A2J7PIN1 A0A1B6H9R4 A0A1B0CIY0 A0A2J7QL19 A0A2H8TCU3 A0A2J7QL26 A0A023EZ96 A0A1B6EU41 A0A1B6M6C7 A0A1B6JJ73 A0A2J7QC67 A0A1B6E8P1 A0A146LN52 A0A224XL30 A0A0A9ZCC5 A0A2J7QC75 T1IC40 A0A069DUA8 A0A2P8XHU0 T1HYD0 A0A2P8Y8I9 A0A224XK01 D4AHX2 A0A1B6M0V4 A0A1B6IEH6 A0A1S3D6K1 A0A1B6GQD0 A0A146KUE8
PDB
4YB9
E-value=1.94798e-11,
Score=168
Ontologies
GO
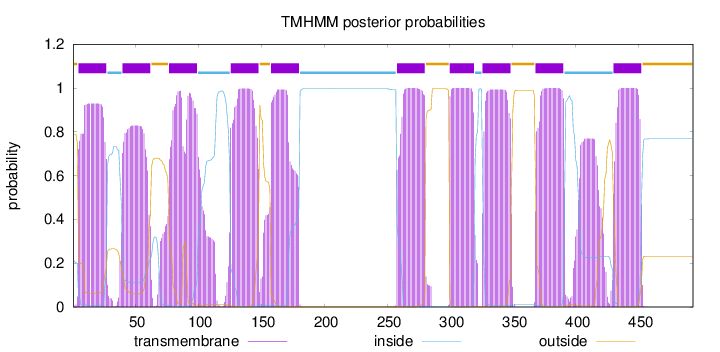
Topology
Length:
493
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
235.98613
Exp number, first 60 AAs:
37.57875
Total prob of N-in:
0.21222
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 27
inside
28 - 39
TMhelix
40 - 62
outside
63 - 76
TMhelix
77 - 99
inside
100 - 125
TMhelix
126 - 148
outside
149 - 157
TMhelix
158 - 180
inside
181 - 257
TMhelix
258 - 280
outside
281 - 299
TMhelix
300 - 319
inside
320 - 325
TMhelix
326 - 348
outside
349 - 367
TMhelix
368 - 390
inside
391 - 429
TMhelix
430 - 452
outside
453 - 493
Population Genetic Test Statistics
Pi
268.417669
Theta
182.923029
Tajima's D
1.432972
CLR
0.184231
CSRT
0.770911454427279
Interpretation
Uncertain
