Gene
KWMTBOMO03901 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010214
Annotation
serine_protease_inhibitor_10_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.018
Sequence
CDS
ATGCGCCAATTGTTTGTGGTTGTGTGCCTAGCAGTGGCCCTTGTCAGTGCACGATGGGTTCGCGAGGGACGTTCCGACCAACCCAGAGACACCAGCTTTATTGGAGATGCTTCCAGTGAACTATCTACAGCTATATTACAGGGTTACATTGATGACAATGAAAACATCGCGTTTTCTCCTCTCGGATACTCTGCTATTTTAGCCATCTTAGCCGAAGGTGCGAAAGGGGAGACCAGAGAACAATTGTATAATGCACTTCATCTGCCCCAAGACCCAGCTTTGACAAGAAAAACATACCAATACATCATGGAGAGACTCAAAAATGTAAACACTTACGCATACAATCAACCGGAGCTCAAGAACTTCTTCTATGTTTACAAAAATTATACAGTCAACGATGAATACAGAAAGATTTTGGAAGAATATTATCTAACAGATGTTAGATCAGTTGAAAAATACAATCCAGAATCTGATGTTCATTCAGACGACGATGACGACGAAGAAGCTATTCGCGTACCGGAGAAGACTGACGAACCCAGTGAGCCTAATGAGGATATGCCAATTAAAACGGAACCAAACGAGAAATTAATATCGTTCGCTTTTGTTGATACCCCAGAAAAGGTAGATGTTACTCAAATTGAATACAAACCGGCAAAGAATATTAAAGAGAAGATTAAGTTAGTGAAAACGTATCCAAAAAAAGAGGACTATACAGATGAAGAGGAAACGATGATTGCTGTAGAAGCTAGGAATCATGCTAGGAGCTTAAATGTAAATAAACCAAATCACGATATATCATCAGGTCTTTCTGTCAACAGCGTCAGCAAGAAGTCTAGTGCGAATAAGTCTAGTTCCGTTATGATCATCTTCAACGGCATGTACTTCCGTGGATCGTGGAAGAAACCATTCGATGTTGTTGAGCCTGGCCTCTTTTATAAATCTAGCTCGGAGAAAAAATCCGTTCAAATGATGAAAGCCACAGGCTCTTTCAATACCGCTTTCATCTCAGCTCTCGACAGCCAAGCCATTTATCTTCCTTATGATGGTGGTCGTTACGCTCTCCTGATTCTCGTCCCGAGGTCTCGTGATGGTCTAACTCGCCTTATTGCTGATCTTCCTTCCGTAACTCTGCAAGACATCGAAGATAGCATGAAGAAGGAGGAACTTCTGCTGTCCATGCCCACGTTCTATGTCGAAACTACCACCAAACCTATTGTTTCATTGGTTAAGTTCGGCGTCAGTAAGATCTTCACACACGATGCAGATTTATCTGGAGTCTCTTCTGACGGAGGTCTGTTCGTTGAAGAGATGGCTCAGCACGTGTCCGTTCGCGTTGACAACTCAGATTCCGTAGCCTCAGAGATGTCAGCTGCCAACGTTGACATGCCCCTTTCGAAGAATTTGCCACTTTCGGAGAAACCAGCCACCAAATTTAGTGTCGACCATCCATTCGTTTTCTTCATCATGGATACTTTAGATAATCTAGTGGTTGTCGCTGGAAAAGTCGTTGACCCAGAAAAACCTAAGCCTTTTGATTTTTGA
Protein
MRQLFVVVCLAVALVSARWVREGRSDQPRDTSFIGDASSELSTAILQGYIDDNENIAFSPLGYSAILAILAEGAKGETREQLYNALHLPQDPALTRKTYQYIMERLKNVNTYAYNQPELKNFFYVYKNYTVNDEYRKILEEYYLTDVRSVEKYNPESDVHSDDDDDEEAIRVPEKTDEPSEPNEDMPIKTEPNEKLISFAFVDTPEKVDVTQIEYKPAKNIKEKIKLVKTYPKKEDYTDEEETMIAVEARNHARSLNVNKPNHDISSGLSVNSVSKKSSANKSSSVMIIFNGMYFRGSWKKPFDVVEPGLFYKSSSEKKSVQMMKATGSFNTAFISALDSQAIYLPYDGGRYALLILVPRSRDGLTRLIADLPSVTLQDIEDSMKKEELLLSMPTFYVETTTKPIVSLVKFGVSKIFTHDADLSGVSSDGGLFVEEMAQHVSVRVDNSDSVASEMSAANVDMPLSKNLPLSEKPATKFSVDHPFVFFIMDTLDNLVVVAGKVVDPEKPKPFDF
Summary
Similarity
Belongs to the serpin family.
Uniprot
Pubmed
EMBL
EU935612
ACG61174.1
BABH01011470
KX117920
APM86798.1
NWSH01000329
+ More
PCG77371.1 KZ150329 PZC71364.1 ODYU01009059 SOQ53203.1 RSAL01000013 RVE53400.1 AK402282 BAM18904.1 KQ460317 KPJ15998.1 AK401042 BAM17664.1 KQ459600 KPI94254.1 KU925764 AOG75389.1 KU925748 AOG75373.1 KU925777 AOG75402.1 KU925723 AOG75348.1 KU925735 AOG75360.1 AGBW02009594 OWR50360.1 GAIX01003395 JAA89165.1 JTDY01003192 KOB69988.1 GAHY01001343 JAA76167.1 GEDC01001061 JAS36237.1 AJWK01020830 CH477279 EAT44983.2 JXUM01090748 KQ563862 KXJ73147.1 CVRI01000014 CRK89704.1 CH479186 EDW39251.1
PCG77371.1 KZ150329 PZC71364.1 ODYU01009059 SOQ53203.1 RSAL01000013 RVE53400.1 AK402282 BAM18904.1 KQ460317 KPJ15998.1 AK401042 BAM17664.1 KQ459600 KPI94254.1 KU925764 AOG75389.1 KU925748 AOG75373.1 KU925777 AOG75402.1 KU925723 AOG75348.1 KU925735 AOG75360.1 AGBW02009594 OWR50360.1 GAIX01003395 JAA89165.1 JTDY01003192 KOB69988.1 GAHY01001343 JAA76167.1 GEDC01001061 JAS36237.1 AJWK01020830 CH477279 EAT44983.2 JXUM01090748 KQ563862 KXJ73147.1 CVRI01000014 CRK89704.1 CH479186 EDW39251.1
Proteomes
Pfam
PF00079 Serpin
Interpro
SUPFAM
SSF56574
SSF56574
Gene 3D
ProteinModelPortal
PDB
3NDA
E-value=3.12707e-18,
Score=226
Ontologies
PANTHER
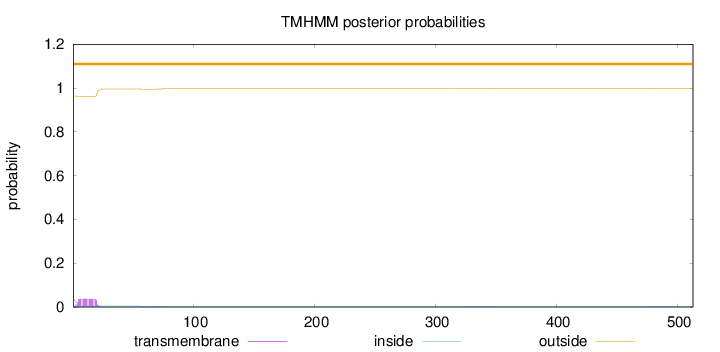
Topology
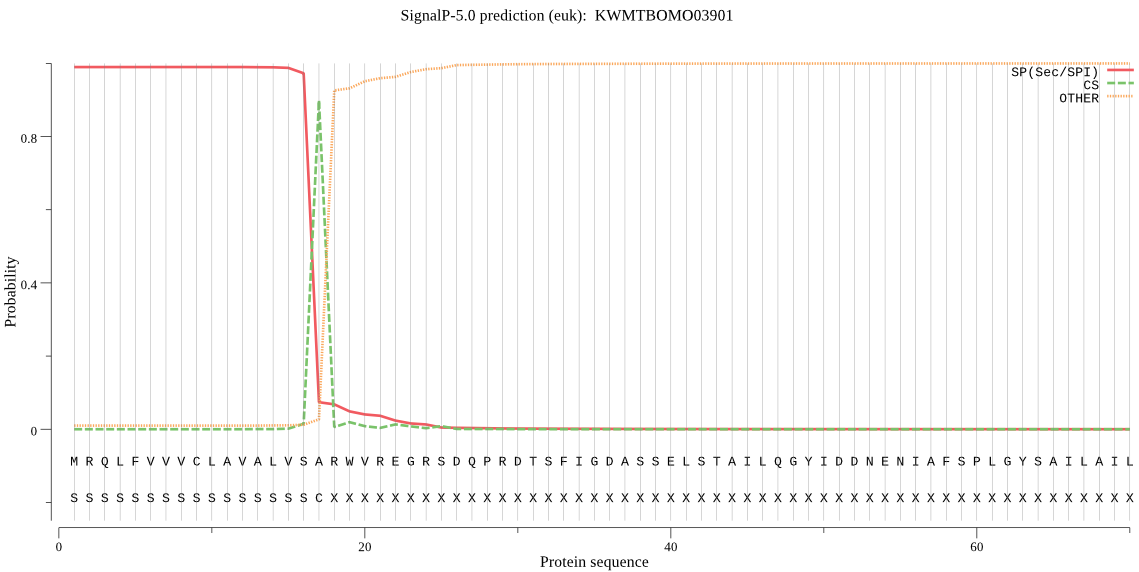
SignalP
Position: 1 - 17,
Likelihood: 0.989061
Length:
513
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.7079
Exp number, first 60 AAs:
0.65032
Total prob of N-in:
0.03591
outside
1 - 513
Population Genetic Test Statistics
Pi
259.637176
Theta
202.073949
Tajima's D
0.192569
CLR
0.983617
CSRT
0.422828858557072
Interpretation
Uncertain
