Gene
KWMTBOMO03900
Pre Gene Modal
BGIBMGA010213
Annotation
serine_protease_inhibitor_11_precursor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.12
Sequence
CDS
ATGCGTGTGTTAATATGTTTTGTCGTAACTTTCGCCAGCCTTCAGACGTTGGATTGTCTTGATACACGAGCACTGTACTCCAGATTGAACTTCTTCGATATTGATTTGCTGAAGTACTCGGCTGAGAATAAACATGGCAATGTGATGGTATCGCCGGCAAGTATAAAATCTACGCTGGCCATGTTGCTGGAAGGAGCTACGGGGGAGACGGCAGACGAAATAAGAAGTGCTTTGAGACTTTCGTCAGTAAAAGACGATTTGAGGGAGCAACTGAATCTTTATTTAAAGGCTTTACATTCAAATTCAACAGAAACCGTTTTAGAAAACGCCAATGCAATATTCGTTTCCAATAAATTAAGACTCAAGAAAGATTACGAGACAAACCTGCGGTATGTGTATTTCTCGGAAGCTGTCCCAGTGAACTTTGGTAATAACGGAGTCGTAGCACAGCAAATAAACGCCTGGGTTAACGGAAAAACCAAAGGACTCATCCCTTCTATTGTAGATCAAGCGCAAATCAACGCTTTCACAGAAATGATTGTGACAAATGCCCTCTACTTCAAGGGTTCTTGGATGCAAGCCTTCAATCCTAAGTTTACACGCCCGGCTTGCTTCTACAACAAGGGAGTTTGCCATAATGTGGCCATGATGGAACTTCAAACAGATATACAATACGCTTATGTGGATAACTTGAGGGCACATGCCGTGGAGCTGCCGTATGCGGGTAAGCGTTACTCTATGATCCTTTTGGTGCCAATAGAACGCGATGGCTGTGCCGCCCTCATCAGAGATCTGCCATTCATGAGTCTACCTCAAATATCCGATCTCCTGGAATCAACTGAAGTTCAGTTGTCCCTGCCCAAATTCACAGTGGACTACAATGAAGACATAATACAAGCTTTGAAAAGTATGCGCATCAACGCCCTTTTCTCACCGTCGGCAAACCTCTCCGGCATCTTTGAAGGCACAACTCCGTATGTTAGCAACCTATATCACAAAGTGCACATGTCAGTGGATGAGCAGGGGACTGTAGCTGCCGCGGCGTCCGCAGCCATGGTTGTGCCGCTCATTAATGATGGCGTAGAACTAAGGGTCGATCGACCCTTCGTTTTCTTCATAAGAGATAATAAGCTTGGCTTGGTGTTATTTGAAGGGAAGATTGACGAGCCCACAGAATATGTTGAGAAGCCCATAAACTATCGAGTTAAAACATCACGACCATTTTTTGGAATCTTTTGA
Protein
MRVLICFVVTFASLQTLDCLDTRALYSRLNFFDIDLLKYSAENKHGNVMVSPASIKSTLAMLLEGATGETADEIRSALRLSSVKDDLREQLNLYLKALHSNSTETVLENANAIFVSNKLRLKKDYETNLRYVYFSEAVPVNFGNNGVVAQQINAWVNGKTKGLIPSIVDQAQINAFTEMIVTNALYFKGSWMQAFNPKFTRPACFYNKGVCHNVAMMELQTDIQYAYVDNLRAHAVELPYAGKRYSMILLVPIERDGCAALIRDLPFMSLPQISDLLESTEVQLSLPKFTVDYNEDIIQALKSMRINALFSPSANLSGIFEGTTPYVSNLYHKVHMSVDEQGTVAAAASAAMVVPLINDGVELRVDRPFVFFIRDNKLGLVLFEGKIDEPTEYVEKPINYRVKTSRPFFGIF
Summary
Similarity
Belongs to the serpin family.
Uniprot
C0J8G1
H9JL12
A0A2A4K156
J7HEW8
A0A2W1BAX2
S4Q008
+ More
X2J6N0 A0A0L7L3E7 A0A194PND8 A0A1E1WTZ3 A0A2H1WMV4 A0A212F9F2 A0A1V1FYH4 A0A2J7QEL8 A0A1Y1L261 A0A067QMH9 A0A1Y1KZL8 A0A1W4XD30 G3XGC5 G3XGC4 A0A0C9RF70 A0A1B6F9W4 A0A1B6I0F2 A0A1B6FDQ0 E0VLN3 E2BHC1 A0A0C9PZQ8 A0A1B6IU83 A0A1B6LMV2 A0A224XK35 A0A2J7QEP5 A0A1B6DK46 A0A1B6DVY9 A0A1B6DL81 K7INJ6 A0A232F3T2 A0A069DZE0 A0A2S2PI23 A0A1B6LQJ3 A0A2K8JMB0 A0A195DGM6 A0A023EZF2 A0A0T6B117 V5HBT7 A0A0K8SVN1 A0A0K8SUU2 A0A0A9Y0L6 A7UI21 E2AL14 A0A146KLK4 A0A195EYZ5 J9JJ46 A7UI32 A0A023GN74 A0A195C2Q5 A0A1E1X361 B0X5Z9 A0A0K8SUF6 A0A0K8SVN5 G3MLB1 Q27085 A0A087SZK7 A0A1Q3FHI8 A0A151X316 A0A0S2CCT2 G3MLE4 A0A2H8TN41 A0A1Q3FH94 A0A023GJN2 A0A195BRF0 A0A224X3W8 F4W5U0 A0A224X3U8 A0A1Q3FH69 A0A1E1XIQ0 A0A158P2S7 A0A0A1X827 A7UI27 A0A0E9Y378 Q75Q63 B4P4R8 A0A023GP80 A0A1E1XJ83 A7UI19 A0A0E9Y1W4 A0A0E9Y315 A0A1E1WWH8 A0A0L0C2U0 W8C425 A0A0E9Y337 A0A0E9Y235 A0A0E9Y2I3 A0A1Q3FHG3 A0A0E9Y268 A0A1F9Q655 A0A3C1JB21
X2J6N0 A0A0L7L3E7 A0A194PND8 A0A1E1WTZ3 A0A2H1WMV4 A0A212F9F2 A0A1V1FYH4 A0A2J7QEL8 A0A1Y1L261 A0A067QMH9 A0A1Y1KZL8 A0A1W4XD30 G3XGC5 G3XGC4 A0A0C9RF70 A0A1B6F9W4 A0A1B6I0F2 A0A1B6FDQ0 E0VLN3 E2BHC1 A0A0C9PZQ8 A0A1B6IU83 A0A1B6LMV2 A0A224XK35 A0A2J7QEP5 A0A1B6DK46 A0A1B6DVY9 A0A1B6DL81 K7INJ6 A0A232F3T2 A0A069DZE0 A0A2S2PI23 A0A1B6LQJ3 A0A2K8JMB0 A0A195DGM6 A0A023EZF2 A0A0T6B117 V5HBT7 A0A0K8SVN1 A0A0K8SUU2 A0A0A9Y0L6 A7UI21 E2AL14 A0A146KLK4 A0A195EYZ5 J9JJ46 A7UI32 A0A023GN74 A0A195C2Q5 A0A1E1X361 B0X5Z9 A0A0K8SUF6 A0A0K8SVN5 G3MLB1 Q27085 A0A087SZK7 A0A1Q3FHI8 A0A151X316 A0A0S2CCT2 G3MLE4 A0A2H8TN41 A0A1Q3FH94 A0A023GJN2 A0A195BRF0 A0A224X3W8 F4W5U0 A0A224X3U8 A0A1Q3FH69 A0A1E1XIQ0 A0A158P2S7 A0A0A1X827 A7UI27 A0A0E9Y378 Q75Q63 B4P4R8 A0A023GP80 A0A1E1XJ83 A7UI19 A0A0E9Y1W4 A0A0E9Y315 A0A1E1WWH8 A0A0L0C2U0 W8C425 A0A0E9Y337 A0A0E9Y235 A0A0E9Y2I3 A0A1Q3FHG3 A0A0E9Y268 A0A1F9Q655 A0A3C1JB21
Pubmed
19150649
19121390
28756777
23622113
26227816
26354079
+ More
22118469 28410430 28004739 24845553 20566863 20798317 20075255 28648823 26334808 25474469 25765539 25401762 17766296 26823975 28503490 22216098 8276848 21719571 21347285 25830018 15652673 17994087 17550304 26108605 24495485 27774985 30148503
22118469 28410430 28004739 24845553 20566863 20798317 20075255 28648823 26334808 25474469 25765539 25401762 17766296 26823975 28503490 22216098 8276848 21719571 21347285 25830018 15652673 17994087 17550304 26108605 24495485 27774985 30148503
EMBL
EU935613
ACG61175.1
BABH01011469
NWSH01000329
PCG77372.1
JN400445
+ More
RSAL01000013 AFQ01144.1 RVE53399.1 KZ150329 PZC71365.1 GAIX01000613 JAA91947.1 KJ127527 AHN49717.1 JTDY01003192 KOB69987.1 KQ459600 KPI94254.1 GDQN01000574 JAT90480.1 ODYU01009759 SOQ54400.1 AGBW02009594 OWR50361.1 FX985306 BAX07319.1 NEVH01015307 PNF27034.1 GEZM01068630 JAV66851.1 KK853245 KDR09407.1 GEZM01068629 JAV66852.1 AB627950 BAL03255.1 AB627949 BAL03254.1 GBYB01007050 JAG76817.1 GECZ01022753 JAS47016.1 GECU01027275 JAS80431.1 GECZ01021676 JAS48093.1 DS235277 EEB14289.1 GL448287 EFN84890.1 GBYB01007043 JAG76810.1 GECU01017217 JAS90489.1 GEBQ01014907 JAT25070.1 GFTR01007923 JAW08503.1 PNF27033.1 GEDC01011261 JAS26037.1 GEDC01007440 JAS29858.1 GEDC01010871 JAS26427.1 NNAY01001103 OXU25130.1 GBGD01000805 JAC88084.1 GGMR01016470 MBY29089.1 GEBQ01014143 JAT25834.1 KY031030 ATU82781.1 KQ980903 KYN11614.1 GBBI01004099 JAC14613.1 LJIG01016282 KRT81068.1 GANP01013810 JAB70658.1 GBRD01008876 JAG56945.1 GBRD01008877 GBRD01008874 GBRD01008870 JAG56951.1 GBHO01018966 GBHO01018965 GBHO01018964 GBHO01018963 JAG24638.1 JAG24639.1 JAG24640.1 JAG24641.1 EU072731 ABS87358.1 GL440429 EFN65867.1 GDHC01021630 JAP96998.1 KQ981905 KYN33351.1 ABLF02039310 ABLF02039313 ABLF02039317 ABLF02039329 EU072742 ABS87369.1 GBBM01000129 JAC35289.1 KQ978379 KYM94473.1 GFAC01005495 JAT93693.1 DS232397 EDS41130.1 GBRD01008875 GBRD01008873 GBRD01008872 JAG56948.1 GBRD01008871 JAG56950.1 JO842662 AEO34279.1 D14483 BAA03374.1 KK112695 KFM58296.1 GFDL01008028 JAV27017.1 KQ982576 KYQ54644.1 KT004437 ALN42752.1 JO842695 AEO34312.1 GFXV01003634 MBW15439.1 GFDL01008105 JAV26940.1 GBBM01001261 JAC34157.1 KQ976417 KYM89580.1 GFBG01000030 JAW07137.1 GL887695 EGI70416.1 GFBG01000031 JAW07136.1 GFDL01008147 JAV26898.1 GFAC01000104 JAT99084.1 ADTU01007439 ADTU01007440 ADTU01007441 GBXI01007412 JAD06880.1 EU072737 ABS87364.1 GAYW01000024 JAI08954.1 AB162827 BAD11156.1 CM000158 EDW91691.1 GBBM01000130 JAC35288.1 GFAC01000107 JAT99081.1 EU072729 ABS87356.1 GAYW01000153 JAI08825.1 GAYW01000142 GAYW01000019 JAI08836.1 GEMQ01000058 JAT91131.1 JRES01000975 KNC26552.1 GAMC01005784 JAC00772.1 GAYW01000139 JAI08839.1 GAYW01000078 JAI08900.1 GAYW01000036 JAI08942.1 GFDL01008070 JAV26975.1 GAYW01000206 JAI08772.1 MGTW01000077 OGR37090.1 DMOH01000040 HAN04937.1
RSAL01000013 AFQ01144.1 RVE53399.1 KZ150329 PZC71365.1 GAIX01000613 JAA91947.1 KJ127527 AHN49717.1 JTDY01003192 KOB69987.1 KQ459600 KPI94254.1 GDQN01000574 JAT90480.1 ODYU01009759 SOQ54400.1 AGBW02009594 OWR50361.1 FX985306 BAX07319.1 NEVH01015307 PNF27034.1 GEZM01068630 JAV66851.1 KK853245 KDR09407.1 GEZM01068629 JAV66852.1 AB627950 BAL03255.1 AB627949 BAL03254.1 GBYB01007050 JAG76817.1 GECZ01022753 JAS47016.1 GECU01027275 JAS80431.1 GECZ01021676 JAS48093.1 DS235277 EEB14289.1 GL448287 EFN84890.1 GBYB01007043 JAG76810.1 GECU01017217 JAS90489.1 GEBQ01014907 JAT25070.1 GFTR01007923 JAW08503.1 PNF27033.1 GEDC01011261 JAS26037.1 GEDC01007440 JAS29858.1 GEDC01010871 JAS26427.1 NNAY01001103 OXU25130.1 GBGD01000805 JAC88084.1 GGMR01016470 MBY29089.1 GEBQ01014143 JAT25834.1 KY031030 ATU82781.1 KQ980903 KYN11614.1 GBBI01004099 JAC14613.1 LJIG01016282 KRT81068.1 GANP01013810 JAB70658.1 GBRD01008876 JAG56945.1 GBRD01008877 GBRD01008874 GBRD01008870 JAG56951.1 GBHO01018966 GBHO01018965 GBHO01018964 GBHO01018963 JAG24638.1 JAG24639.1 JAG24640.1 JAG24641.1 EU072731 ABS87358.1 GL440429 EFN65867.1 GDHC01021630 JAP96998.1 KQ981905 KYN33351.1 ABLF02039310 ABLF02039313 ABLF02039317 ABLF02039329 EU072742 ABS87369.1 GBBM01000129 JAC35289.1 KQ978379 KYM94473.1 GFAC01005495 JAT93693.1 DS232397 EDS41130.1 GBRD01008875 GBRD01008873 GBRD01008872 JAG56948.1 GBRD01008871 JAG56950.1 JO842662 AEO34279.1 D14483 BAA03374.1 KK112695 KFM58296.1 GFDL01008028 JAV27017.1 KQ982576 KYQ54644.1 KT004437 ALN42752.1 JO842695 AEO34312.1 GFXV01003634 MBW15439.1 GFDL01008105 JAV26940.1 GBBM01001261 JAC34157.1 KQ976417 KYM89580.1 GFBG01000030 JAW07137.1 GL887695 EGI70416.1 GFBG01000031 JAW07136.1 GFDL01008147 JAV26898.1 GFAC01000104 JAT99084.1 ADTU01007439 ADTU01007440 ADTU01007441 GBXI01007412 JAD06880.1 EU072737 ABS87364.1 GAYW01000024 JAI08954.1 AB162827 BAD11156.1 CM000158 EDW91691.1 GBBM01000130 JAC35288.1 GFAC01000107 JAT99081.1 EU072729 ABS87356.1 GAYW01000153 JAI08825.1 GAYW01000142 GAYW01000019 JAI08836.1 GEMQ01000058 JAT91131.1 JRES01000975 KNC26552.1 GAMC01005784 JAC00772.1 GAYW01000139 JAI08839.1 GAYW01000078 JAI08900.1 GAYW01000036 JAI08942.1 GFDL01008070 JAV26975.1 GAYW01000206 JAI08772.1 MGTW01000077 OGR37090.1 DMOH01000040 HAN04937.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000053268
UP000007151
+ More
UP000235965 UP000027135 UP000192223 UP000009046 UP000008237 UP000002358 UP000215335 UP000078492 UP000000311 UP000078541 UP000007819 UP000078542 UP000002320 UP000054359 UP000075809 UP000078540 UP000007755 UP000005205 UP000002282 UP000037069
UP000235965 UP000027135 UP000192223 UP000009046 UP000008237 UP000002358 UP000215335 UP000078492 UP000000311 UP000078541 UP000007819 UP000078542 UP000002320 UP000054359 UP000075809 UP000078540 UP000007755 UP000005205 UP000002282 UP000037069
Pfam
PF00079 Serpin
Interpro
SUPFAM
SSF56574
SSF56574
Gene 3D
ProteinModelPortal
C0J8G1
H9JL12
A0A2A4K156
J7HEW8
A0A2W1BAX2
S4Q008
+ More
X2J6N0 A0A0L7L3E7 A0A194PND8 A0A1E1WTZ3 A0A2H1WMV4 A0A212F9F2 A0A1V1FYH4 A0A2J7QEL8 A0A1Y1L261 A0A067QMH9 A0A1Y1KZL8 A0A1W4XD30 G3XGC5 G3XGC4 A0A0C9RF70 A0A1B6F9W4 A0A1B6I0F2 A0A1B6FDQ0 E0VLN3 E2BHC1 A0A0C9PZQ8 A0A1B6IU83 A0A1B6LMV2 A0A224XK35 A0A2J7QEP5 A0A1B6DK46 A0A1B6DVY9 A0A1B6DL81 K7INJ6 A0A232F3T2 A0A069DZE0 A0A2S2PI23 A0A1B6LQJ3 A0A2K8JMB0 A0A195DGM6 A0A023EZF2 A0A0T6B117 V5HBT7 A0A0K8SVN1 A0A0K8SUU2 A0A0A9Y0L6 A7UI21 E2AL14 A0A146KLK4 A0A195EYZ5 J9JJ46 A7UI32 A0A023GN74 A0A195C2Q5 A0A1E1X361 B0X5Z9 A0A0K8SUF6 A0A0K8SVN5 G3MLB1 Q27085 A0A087SZK7 A0A1Q3FHI8 A0A151X316 A0A0S2CCT2 G3MLE4 A0A2H8TN41 A0A1Q3FH94 A0A023GJN2 A0A195BRF0 A0A224X3W8 F4W5U0 A0A224X3U8 A0A1Q3FH69 A0A1E1XIQ0 A0A158P2S7 A0A0A1X827 A7UI27 A0A0E9Y378 Q75Q63 B4P4R8 A0A023GP80 A0A1E1XJ83 A7UI19 A0A0E9Y1W4 A0A0E9Y315 A0A1E1WWH8 A0A0L0C2U0 W8C425 A0A0E9Y337 A0A0E9Y235 A0A0E9Y2I3 A0A1Q3FHG3 A0A0E9Y268 A0A1F9Q655 A0A3C1JB21
X2J6N0 A0A0L7L3E7 A0A194PND8 A0A1E1WTZ3 A0A2H1WMV4 A0A212F9F2 A0A1V1FYH4 A0A2J7QEL8 A0A1Y1L261 A0A067QMH9 A0A1Y1KZL8 A0A1W4XD30 G3XGC5 G3XGC4 A0A0C9RF70 A0A1B6F9W4 A0A1B6I0F2 A0A1B6FDQ0 E0VLN3 E2BHC1 A0A0C9PZQ8 A0A1B6IU83 A0A1B6LMV2 A0A224XK35 A0A2J7QEP5 A0A1B6DK46 A0A1B6DVY9 A0A1B6DL81 K7INJ6 A0A232F3T2 A0A069DZE0 A0A2S2PI23 A0A1B6LQJ3 A0A2K8JMB0 A0A195DGM6 A0A023EZF2 A0A0T6B117 V5HBT7 A0A0K8SVN1 A0A0K8SUU2 A0A0A9Y0L6 A7UI21 E2AL14 A0A146KLK4 A0A195EYZ5 J9JJ46 A7UI32 A0A023GN74 A0A195C2Q5 A0A1E1X361 B0X5Z9 A0A0K8SUF6 A0A0K8SVN5 G3MLB1 Q27085 A0A087SZK7 A0A1Q3FHI8 A0A151X316 A0A0S2CCT2 G3MLE4 A0A2H8TN41 A0A1Q3FH94 A0A023GJN2 A0A195BRF0 A0A224X3W8 F4W5U0 A0A224X3U8 A0A1Q3FH69 A0A1E1XIQ0 A0A158P2S7 A0A0A1X827 A7UI27 A0A0E9Y378 Q75Q63 B4P4R8 A0A023GP80 A0A1E1XJ83 A7UI19 A0A0E9Y1W4 A0A0E9Y315 A0A1E1WWH8 A0A0L0C2U0 W8C425 A0A0E9Y337 A0A0E9Y235 A0A0E9Y2I3 A0A1Q3FHG3 A0A0E9Y268 A0A1F9Q655 A0A3C1JB21
PDB
5CDZ
E-value=1.56199e-42,
Score=435
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.976467
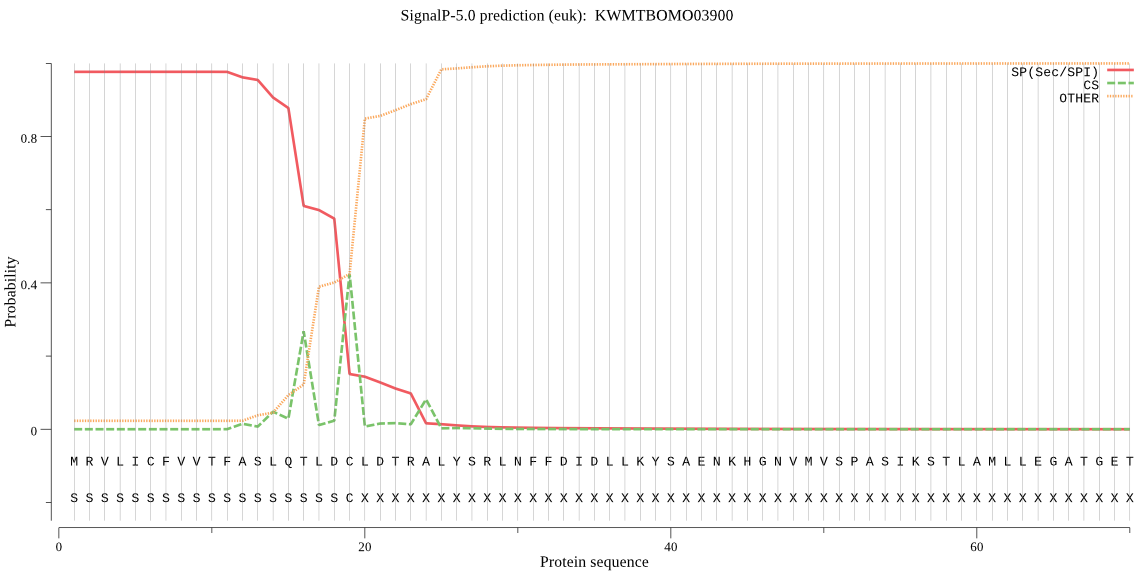
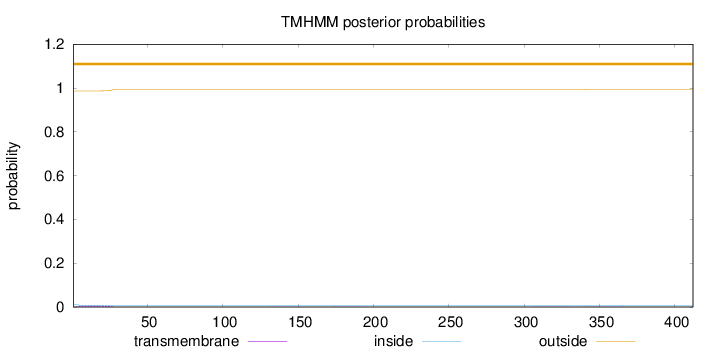
Length:
412
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2037
Exp number, first 60 AAs:
0.16766
Total prob of N-in:
0.01363
outside
1 - 412
Population Genetic Test Statistics
Pi
315.086913
Theta
187.310743
Tajima's D
1.470407
CLR
0.626802
CSRT
0.781810909454527
Interpretation
Uncertain
