Pre Gene Modal
BGIBMGA010209
Annotation
cell_differentiation_protein_RCD1_homolog_[Bombyx_mori]
Full name
CCR4-NOT transcription complex subunit 9
Alternative Name
Cell differentiation protein RQCD1 homolog
Location in the cell
PlasmaMembrane Reliability : 2.732
Sequence
CDS
ATGAGTCGTTTAACGCCAATCGTACAAAATATGATTAATATGAGCGCACAGCAGAGTCCGGCAAATATGCAGGCTGCCGTGGACAGGGAAAAGATATATTCCTGGATTCTAGAGCTATGCAATCCCGACACTCGTGAAAATGCGCTATTAGAACTGAGTAAAAAACGAGAAGTGGTTCCTGACTTGGCGCCAATGCTGTGGCATAGCTTCGGTACAATCGCTGCTCTCCTACAGGAAATAACCAACATATACGTGGCAATGATTCCTCCGACTTTGACTGCTCATCAGAGCAACCGTGTCTGTAATGCTTTGGCTTTGATGCAATGTGTTGCATCTCATCCAGAAACAAGATCTGCGTTTCTTCAAGCCCATGTTCCCTTGTTCCTTTACCCATTCCTCCACACTGTTTCTAAAACTAGACCTTTTGAATACCTACGTCTTACCAGTCTTGGAGTAATAGGTGCATTAGTGAAAACGGATGAGCAAGAAGTTATAACATTTCTATTGACTACCGAAATTATACCACTGTGTCTGCGCATTATGGAAAATGGATCTGAACTCTCTAAAACTGTAGCTACGTTTATCCTACAGAAAATTTTGCTTGACGACAGTGGACTTTGTTATATTTGCCAAACCTATGACAGATTTTCTCATGTTGCCATGATTTTGGGGAAAATGGTACTTTCACTAGCTAAAGATCCATCAGCTCGGTTGTTAAAGCATGTAGTAAGATGTTACCTTCGTTTATCTGACAACCCAAGAGCAAGAGAAGCTTTAAGACAATGCTTGCCAGACCAACTCCGTGATGCAACTTTTACAGCATGCCTTCAAGAAGACAATTCTACAAAGCATTGGTTGGCTCAACTCATTAAGAACTTGGAAGCACAGCCTCCTCCGGCCAGTCCAGCTCAGCAGAACATGTGA
Protein
MSRLTPIVQNMINMSAQQSPANMQAAVDREKIYSWILELCNPDTRENALLELSKKREVVPDLAPMLWHSFGTIAALLQEITNIYVAMIPPTLTAHQSNRVCNALALMQCVASHPETRSAFLQAHVPLFLYPFLHTVSKTRPFEYLRLTSLGVIGALVKTDEQEVITFLLTTEIIPLCLRIMENGSELSKTVATFILQKILLDDSGLCYICQTYDRFSHVAMILGKMVLSLAKDPSARLLKHVVRCYLRLSDNPRAREALRQCLPDQLRDATFTACLQEDNSTKHWLAQLIKNLEAQPPPASPAQQNM
Summary
Description
Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Involved in down-regulation of MYB- and JUN-dependent transcription. Enhances ligand-dependent transcriptional activity of nuclear hormone receptors. May play a role in cell differentiation.
Subunit
Homodimer. Component of the CCR4-NOT complex.
Homodimer. Component of the CCR4-NOT complex (By similarity).
Homodimer. Component of the CCR4-NOT complex (By similarity).
Similarity
Belongs to the CNOT9 family.
Keywords
Activator
Complete proteome
Cytoplasm
Nucleus
Reference proteome
Repressor
RNA-mediated gene silencing
Transcription
Transcription regulation
Translation regulation
Feature
chain CCR4-NOT transcription complex subunit 9
Uniprot
A0A212ERZ8
S4PBB0
A0A0L7LK19
A0A2A4JZC0
I4DNJ6
A0A194RPI8
+ More
A0A194Q5R5 H9JL08 D4NXP4 A0A2H1WMZ9 A0A222AIZ0 A0A3S2NHH9 A0A2J7PDY7 A0A1B6EI93 A0A1B6EIN7 A0A1B6LJZ7 E0VHW2 A0A2A3E496 A0A088AH98 A0A1B6CYZ2 A0A1B6E3Y4 A0A0J7K982 A0A195DCY9 A0A0N0BBH7 A0A195FW70 E2BD53 A0A158NS60 E2AWG5 A0A151WH26 F4WJQ1 A0A1W4W5G8 A0A232F2M2 E9IR20 A0A0P4VMN4 R4FLK5 A0A2R7VTL1 A0A0V0G8H5 A0A026X1J9 A0A023F9H4 A0A161TH70 A0A224XGR6 A0A069DR60 D6WJX4 A0A1Y1NL77 J3JWL2 A0A023EQ69 Q174K1 A0A195BC81 A0A0A9YV04 A0A1B6IXH7 A0A146LPW8 B0WBV0 A0A1Q3F885 A0A0K8TMC6 A0A1S3HMC2 A0A1S3CXA5 A0A1S3CYL7 A0A336MA08 A0A1Q3F8B8 A0A1Q3F867 A0A1L8EVQ1 Q6P819 Q6IP65 A0A091FJB6 A0A091PCG2 E9HFR5 A0A2I0UKJ1 H3BDT3 A0A091NMQ8 A0A2Y9R5U6 A0A164L5M9 A0A3Q0CQS0 A0A093E6U2 A0A093JBC6 A0A093FE01 A0A093Q7M1 A0A091U6L4 A0A091I7G3 A0A091UUS7 A0A091M981 A0A087V2K0 A0A091JNW0 A0A099ZZ37 A0A091UMR0 A0A093P5Z6 A0A091SVA7 A0A093J034 A0A091EAI7 A0A091RP85 A0A091T2F1 A0A091P204 A0A087QTT3 A0A094L647 A0A091QAC3 R0LIK7 A0A091N894 H0Z510 A0A2K5HP21 U6DA15 K9IID3
A0A194Q5R5 H9JL08 D4NXP4 A0A2H1WMZ9 A0A222AIZ0 A0A3S2NHH9 A0A2J7PDY7 A0A1B6EI93 A0A1B6EIN7 A0A1B6LJZ7 E0VHW2 A0A2A3E496 A0A088AH98 A0A1B6CYZ2 A0A1B6E3Y4 A0A0J7K982 A0A195DCY9 A0A0N0BBH7 A0A195FW70 E2BD53 A0A158NS60 E2AWG5 A0A151WH26 F4WJQ1 A0A1W4W5G8 A0A232F2M2 E9IR20 A0A0P4VMN4 R4FLK5 A0A2R7VTL1 A0A0V0G8H5 A0A026X1J9 A0A023F9H4 A0A161TH70 A0A224XGR6 A0A069DR60 D6WJX4 A0A1Y1NL77 J3JWL2 A0A023EQ69 Q174K1 A0A195BC81 A0A0A9YV04 A0A1B6IXH7 A0A146LPW8 B0WBV0 A0A1Q3F885 A0A0K8TMC6 A0A1S3HMC2 A0A1S3CXA5 A0A1S3CYL7 A0A336MA08 A0A1Q3F8B8 A0A1Q3F867 A0A1L8EVQ1 Q6P819 Q6IP65 A0A091FJB6 A0A091PCG2 E9HFR5 A0A2I0UKJ1 H3BDT3 A0A091NMQ8 A0A2Y9R5U6 A0A164L5M9 A0A3Q0CQS0 A0A093E6U2 A0A093JBC6 A0A093FE01 A0A093Q7M1 A0A091U6L4 A0A091I7G3 A0A091UUS7 A0A091M981 A0A087V2K0 A0A091JNW0 A0A099ZZ37 A0A091UMR0 A0A093P5Z6 A0A091SVA7 A0A093J034 A0A091EAI7 A0A091RP85 A0A091T2F1 A0A091P204 A0A087QTT3 A0A094L647 A0A091QAC3 R0LIK7 A0A091N894 H0Z510 A0A2K5HP21 U6DA15 K9IID3
Pubmed
EMBL
AGBW02012891
OWR44270.1
GAIX01006002
JAA86558.1
JTDY01000889
KOB75546.1
+ More
NWSH01000329 PCG77365.1 AK402971 BAM19486.1 KQ460075 KPJ17936.1 KQ459582 KPI98740.1 BABH01011457 GU454861 ADD54617.1 ODYU01009759 SOQ54408.1 KY563841 ASO76347.1 RSAL01003985 RVE40203.1 NEVH01026122 PNF14545.1 GECZ01032112 JAS37657.1 GECZ01031980 JAS37789.1 GEBQ01015977 JAT24000.1 DS235172 EEB12968.1 KZ288427 PBC25901.1 GEDC01018835 JAS18463.1 GEDC01004652 JAS32646.1 LBMM01011538 KMQ86776.1 KQ980989 KYN10746.1 KQ438551 KOX67191.1 KQ981208 KYN44688.1 GL447569 EFN86361.1 ADTU01024525 ADTU01024526 GL443286 EFN62265.1 KQ983136 KYQ47117.1 GL888186 EGI65543.1 NNAY01001156 OXU24945.1 GL765043 EFZ16974.1 GDKW01002478 JAI54117.1 GAHY01002074 JAA75436.1 KK854081 PTY10864.1 GECL01001877 JAP04247.1 KK107031 QOIP01000008 EZA62127.1 RLU19286.1 RLU19625.1 GBBI01000561 JAC18151.1 GEMB01000704 JAS02432.1 GFTR01004740 JAW11686.1 GBGD01002331 JAC86558.1 KQ971342 EFA03649.1 GEZM01003813 GEZM01003812 JAV96547.1 BT127630 AEE62592.1 GAPW01002904 JAC10694.1 CH477409 EAT41504.1 KQ976528 KYM81827.1 GBHO01006747 GBRD01003236 JAG36857.1 JAG62585.1 GECU01016059 JAS91647.1 GDHC01009300 JAQ09329.1 DS231882 EDS42880.1 GFDL01011322 JAV23723.1 GDAI01002300 JAI15303.1 UFQS01000552 UFQT01000552 SSX04844.1 SSX25207.1 GFDL01011307 JAV23738.1 GFDL01011275 JAV23770.1 CM004483 CM004482 OCT61020.1 OCT63424.1 CR760361 BC061412 BC072053 KL447081 KFO69703.1 KK670975 KFQ04943.1 GL732637 EFX69422.1 KZ505705 PKU46560.1 AFYH01036142 AFYH01036143 KK837592 KFP80780.1 LRGB01003193 KZS03816.1 KL457336 KFV10166.1 KK611209 KFW12335.1 KK631483 KFV55937.1 KL670757 KFW80157.1 KK419073 KFQ86096.1 KL218243 KFP03320.1 KL410197 KFQ94719.1 KK525734 KFP68056.1 KL477547 KFO06842.1 KK502214 KFP21623.1 KL869907 KGL86403.1 KK458191 KFQ77619.1 KL225119 KFW67652.1 KK935416 KFQ46922.1 KK567526 KFW05247.1 KK718211 KFO55053.1 KK822712 KFQ40759.1 KK496958 KFQ66114.1 KK649309 KFQ00853.1 KL225902 KFM04637.1 KL277532 KFZ66823.1 KK692832 KFQ22562.1 KB743225 EOB00198.1 KL379101 KFP86038.1 ABQF01010787 HAAF01007632 CCP79456.1 GABZ01007202 JAA46323.1
NWSH01000329 PCG77365.1 AK402971 BAM19486.1 KQ460075 KPJ17936.1 KQ459582 KPI98740.1 BABH01011457 GU454861 ADD54617.1 ODYU01009759 SOQ54408.1 KY563841 ASO76347.1 RSAL01003985 RVE40203.1 NEVH01026122 PNF14545.1 GECZ01032112 JAS37657.1 GECZ01031980 JAS37789.1 GEBQ01015977 JAT24000.1 DS235172 EEB12968.1 KZ288427 PBC25901.1 GEDC01018835 JAS18463.1 GEDC01004652 JAS32646.1 LBMM01011538 KMQ86776.1 KQ980989 KYN10746.1 KQ438551 KOX67191.1 KQ981208 KYN44688.1 GL447569 EFN86361.1 ADTU01024525 ADTU01024526 GL443286 EFN62265.1 KQ983136 KYQ47117.1 GL888186 EGI65543.1 NNAY01001156 OXU24945.1 GL765043 EFZ16974.1 GDKW01002478 JAI54117.1 GAHY01002074 JAA75436.1 KK854081 PTY10864.1 GECL01001877 JAP04247.1 KK107031 QOIP01000008 EZA62127.1 RLU19286.1 RLU19625.1 GBBI01000561 JAC18151.1 GEMB01000704 JAS02432.1 GFTR01004740 JAW11686.1 GBGD01002331 JAC86558.1 KQ971342 EFA03649.1 GEZM01003813 GEZM01003812 JAV96547.1 BT127630 AEE62592.1 GAPW01002904 JAC10694.1 CH477409 EAT41504.1 KQ976528 KYM81827.1 GBHO01006747 GBRD01003236 JAG36857.1 JAG62585.1 GECU01016059 JAS91647.1 GDHC01009300 JAQ09329.1 DS231882 EDS42880.1 GFDL01011322 JAV23723.1 GDAI01002300 JAI15303.1 UFQS01000552 UFQT01000552 SSX04844.1 SSX25207.1 GFDL01011307 JAV23738.1 GFDL01011275 JAV23770.1 CM004483 CM004482 OCT61020.1 OCT63424.1 CR760361 BC061412 BC072053 KL447081 KFO69703.1 KK670975 KFQ04943.1 GL732637 EFX69422.1 KZ505705 PKU46560.1 AFYH01036142 AFYH01036143 KK837592 KFP80780.1 LRGB01003193 KZS03816.1 KL457336 KFV10166.1 KK611209 KFW12335.1 KK631483 KFV55937.1 KL670757 KFW80157.1 KK419073 KFQ86096.1 KL218243 KFP03320.1 KL410197 KFQ94719.1 KK525734 KFP68056.1 KL477547 KFO06842.1 KK502214 KFP21623.1 KL869907 KGL86403.1 KK458191 KFQ77619.1 KL225119 KFW67652.1 KK935416 KFQ46922.1 KK567526 KFW05247.1 KK718211 KFO55053.1 KK822712 KFQ40759.1 KK496958 KFQ66114.1 KK649309 KFQ00853.1 KL225902 KFM04637.1 KL277532 KFZ66823.1 KK692832 KFQ22562.1 KB743225 EOB00198.1 KL379101 KFP86038.1 ABQF01010787 HAAF01007632 CCP79456.1 GABZ01007202 JAA46323.1
Proteomes
UP000007151
UP000037510
UP000218220
UP000053240
UP000053268
UP000005204
+ More
UP000283053 UP000235965 UP000009046 UP000242457 UP000005203 UP000036403 UP000078492 UP000053105 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000007755 UP000192223 UP000215335 UP000053097 UP000279307 UP000007266 UP000008820 UP000078540 UP000002320 UP000085678 UP000079169 UP000186698 UP000008143 UP000053760 UP000000305 UP000008672 UP000248480 UP000076858 UP000189706 UP000053258 UP000054308 UP000053283 UP000053119 UP000053858 UP000054081 UP000052976 UP000053286 UP000007754 UP000233080
UP000283053 UP000235965 UP000009046 UP000242457 UP000005203 UP000036403 UP000078492 UP000053105 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000007755 UP000192223 UP000215335 UP000053097 UP000279307 UP000007266 UP000008820 UP000078540 UP000002320 UP000085678 UP000079169 UP000186698 UP000008143 UP000053760 UP000000305 UP000008672 UP000248480 UP000076858 UP000189706 UP000053258 UP000054308 UP000053283 UP000053119 UP000053858 UP000054081 UP000052976 UP000053286 UP000007754 UP000233080
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A212ERZ8
S4PBB0
A0A0L7LK19
A0A2A4JZC0
I4DNJ6
A0A194RPI8
+ More
A0A194Q5R5 H9JL08 D4NXP4 A0A2H1WMZ9 A0A222AIZ0 A0A3S2NHH9 A0A2J7PDY7 A0A1B6EI93 A0A1B6EIN7 A0A1B6LJZ7 E0VHW2 A0A2A3E496 A0A088AH98 A0A1B6CYZ2 A0A1B6E3Y4 A0A0J7K982 A0A195DCY9 A0A0N0BBH7 A0A195FW70 E2BD53 A0A158NS60 E2AWG5 A0A151WH26 F4WJQ1 A0A1W4W5G8 A0A232F2M2 E9IR20 A0A0P4VMN4 R4FLK5 A0A2R7VTL1 A0A0V0G8H5 A0A026X1J9 A0A023F9H4 A0A161TH70 A0A224XGR6 A0A069DR60 D6WJX4 A0A1Y1NL77 J3JWL2 A0A023EQ69 Q174K1 A0A195BC81 A0A0A9YV04 A0A1B6IXH7 A0A146LPW8 B0WBV0 A0A1Q3F885 A0A0K8TMC6 A0A1S3HMC2 A0A1S3CXA5 A0A1S3CYL7 A0A336MA08 A0A1Q3F8B8 A0A1Q3F867 A0A1L8EVQ1 Q6P819 Q6IP65 A0A091FJB6 A0A091PCG2 E9HFR5 A0A2I0UKJ1 H3BDT3 A0A091NMQ8 A0A2Y9R5U6 A0A164L5M9 A0A3Q0CQS0 A0A093E6U2 A0A093JBC6 A0A093FE01 A0A093Q7M1 A0A091U6L4 A0A091I7G3 A0A091UUS7 A0A091M981 A0A087V2K0 A0A091JNW0 A0A099ZZ37 A0A091UMR0 A0A093P5Z6 A0A091SVA7 A0A093J034 A0A091EAI7 A0A091RP85 A0A091T2F1 A0A091P204 A0A087QTT3 A0A094L647 A0A091QAC3 R0LIK7 A0A091N894 H0Z510 A0A2K5HP21 U6DA15 K9IID3
A0A194Q5R5 H9JL08 D4NXP4 A0A2H1WMZ9 A0A222AIZ0 A0A3S2NHH9 A0A2J7PDY7 A0A1B6EI93 A0A1B6EIN7 A0A1B6LJZ7 E0VHW2 A0A2A3E496 A0A088AH98 A0A1B6CYZ2 A0A1B6E3Y4 A0A0J7K982 A0A195DCY9 A0A0N0BBH7 A0A195FW70 E2BD53 A0A158NS60 E2AWG5 A0A151WH26 F4WJQ1 A0A1W4W5G8 A0A232F2M2 E9IR20 A0A0P4VMN4 R4FLK5 A0A2R7VTL1 A0A0V0G8H5 A0A026X1J9 A0A023F9H4 A0A161TH70 A0A224XGR6 A0A069DR60 D6WJX4 A0A1Y1NL77 J3JWL2 A0A023EQ69 Q174K1 A0A195BC81 A0A0A9YV04 A0A1B6IXH7 A0A146LPW8 B0WBV0 A0A1Q3F885 A0A0K8TMC6 A0A1S3HMC2 A0A1S3CXA5 A0A1S3CYL7 A0A336MA08 A0A1Q3F8B8 A0A1Q3F867 A0A1L8EVQ1 Q6P819 Q6IP65 A0A091FJB6 A0A091PCG2 E9HFR5 A0A2I0UKJ1 H3BDT3 A0A091NMQ8 A0A2Y9R5U6 A0A164L5M9 A0A3Q0CQS0 A0A093E6U2 A0A093JBC6 A0A093FE01 A0A093Q7M1 A0A091U6L4 A0A091I7G3 A0A091UUS7 A0A091M981 A0A087V2K0 A0A091JNW0 A0A099ZZ37 A0A091UMR0 A0A093P5Z6 A0A091SVA7 A0A093J034 A0A091EAI7 A0A091RP85 A0A091T2F1 A0A091P204 A0A087QTT3 A0A094L647 A0A091QAC3 R0LIK7 A0A091N894 H0Z510 A0A2K5HP21 U6DA15 K9IID3
PDB
4CT6
E-value=8.00519e-131,
Score=1195
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
P-body
Cytoplasm
P-body
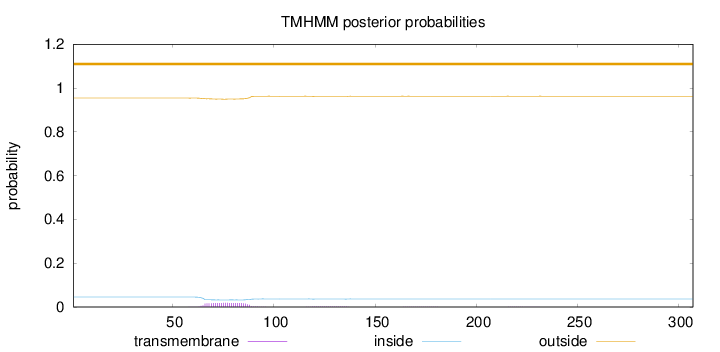
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.458919999999999
Exp number, first 60 AAs:
0.00128
Total prob of N-in:
0.04570
outside
1 - 307
Population Genetic Test Statistics
Pi
138.205203
Theta
129.440259
Tajima's D
0.308598
CLR
4.617438
CSRT
0.458977051147443
Interpretation
Uncertain
