Pre Gene Modal
BGIBMGA010165
Annotation
PREDICTED:_HEAT_repeat-containing_protein_5B_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.048
Sequence
CDS
ATGGAAGTGTCCCATTCCTTGACTCTAAACGAAACAGCATTGCAGCAGCTGCCAGATGATAAAAAGCCACATTTCATATTTGAATGGCTACGGTTCCTTGACAAGGTGTTGGTTGCAGCACACAAGAATGATATTAAAGCTTGCCAACAAAAACTTGTTGCCCAATTAGTTAATCTTCTACGTGAATCCCTTGGTGCACCTGCTAGAAAACTGCTTGCCCGGAATTTAGCCATACTCTTCTCTGTCGGGGATACCTTCCTTTTATTTGATACAGTAAATAAGTGCAATGACATCATCAAAGTGAAGGATGACTCGCCTTCATACATGCCGACTAGATTGGCTGCAATATGCTGCCTTGGTAGTATGTATGAGAAGCTAGGTAGAATGATGGGAAGATCACATGAAGAAACTGTTACTACTCTGGTCAGGTCCTTAAAGAGTGCAGAATCGCAAACCAGGCTTGAAATCATGAATACACTGGAAAAGATCTGTGTTGGGATAGGATCTGCTGCTCCAGCGGCAGTGAGAGAAGTATCTCGTGCTGCTCGTACAGCATTGATGTCAGATCGGTCTGCAGCAGTGAGAGCTGCGGCAGCACACACCCTGACTAACTTGTTGCCACTAATGAGACTGACACCAGCAGATCTTGATGCTATAGCTGCTGCTTGTCTCCGTGCCGCCAATCCCAGTGATTATATTTTAAGATGTGCAATTGCGGAACTTTTGGGATCTCTAATAGCAACAACACAAGCTGCTGAAGTGACGAATAACAACAAGTTTAAAGTCGGCTTGCAACCAGTGCAACAAAATAAGAAGGATCCTCCTAAAAATGTAGTCTCATTAGACGAAGCTCTGAACTTATTGATGGGCGCTTTTCTAAGAGGGGGCACGTCGTTCCTAAAGGGTGAAATTATTAAATCTGGTTCCGGTGTCAACAGGGAAGTTAGAGTATGTGTTACAAATGCATATGTCGTGTTCGTGCAGAACATGGGAGGAGTTTGGTTGGAACGTAACATAACTACGTTTTTAGCTCACGTATTGGATTTGGTGGCTAATCCTAAAGCAGCTAGTTCTCATGTCGACGCGGTTTATTCAAGAAAGTGCATTAGTTATATTCTGCGCAACACTCTCGGCAAGATGCTAGGGGAGAAAGCACAAGCGTCGGCGTGTCGGGAGATCATACAGATTGTTGTCAAACAAATGAACAGCATCGACTTTAATCCGGAAAACGCGAAGGATTGTAATCAGGAAACTTTGTTTAGTCAGCACCTTTTAGTGTGTGCTCTAATGGAAGCGGGAGAATTGGCCTTGCAGCTCGGTACTTCAATTCAGAATCTCATATCAGATACATCATTGAATATGGTTGAAGCTATATGTAGTGTGAGTGAATTGGAAGTGCCTCAAAAATCAACGCAAAAAATGTCGATTTTTGATGTTGTTTTCCATTGTTGA
Protein
MEVSHSLTLNETALQQLPDDKKPHFIFEWLRFLDKVLVAAHKNDIKACQQKLVAQLVNLLRESLGAPARKLLARNLAILFSVGDTFLLFDTVNKCNDIIKVKDDSPSYMPTRLAAICCLGSMYEKLGRMMGRSHEETVTTLVRSLKSAESQTRLEIMNTLEKICVGIGSAAPAAVREVSRAARTALMSDRSAAVRAAAAHTLTNLLPLMRLTPADLDAIAAACLRAANPSDYILRCAIAELLGSLIATTQAAEVTNNNKFKVGLQPVQQNKKDPPKNVVSLDEALNLLMGAFLRGGTSFLKGEIIKSGSGVNREVRVCVTNAYVVFVQNMGGVWLERNITTFLAHVLDLVANPKAASSHVDAVYSRKCISYILRNTLGKMLGEKAQASACREIIQIVVKQMNSIDFNPENAKDCNQETLFSQHLLVCALMEAGELALQLGTSIQNLISDTSLNMVEAICSVSELEVPQKSTQKMSIFDVVFHC
Summary
Uniprot
H9JKW4
A0A2A4K015
A0A2H1WMW5
A0A3S2NZW0
A0A194RKS7
A0A2W1C1K1
+ More
A0A067QJA9 A0A2M3YZX2 A0A0P6ISX1 A0A1S4FV30 Q16N63 A0A182K0E0 A0A088AD24 B4R0D3 A0A1B6DSD1 A0A139WP21 D6W798 A0A182LQK5 Q7PEZ3 A0A182NMS5 A0A182HZJ8 A0A182UG76 A0A182GHQ0 A0A182X364 A0A182NZG0 A0A182IL48 U4UQA3 A0A182QPM0 W4VS74 A0A1Y1LQW1 A0A084W110 T1PP11 A0A1W4WFD5 E0VLY1 A0A1B6GN91 A0A0C9Q9W5 A0A0C9RRK4 A0A1B6G0A3 A0A0C9RDI9 A0A232FDY5 A0A1Q3FIR7 A0A1Q3FIE0 B0WFJ4 A0A1J1IIL2 A0A2A3EKW7 K7J8E1 A0A195AUF0 Q1WWE5 A0A3L8DH26 A0A158NN99 E9IVN3 A0A195FWC2 A0A0M8ZUQ0 A0A0J7NJK1 A0A195CT14 E2A542 E2C319 A0A1B0GQM9 A0A026WSX8 A0A151XA78 A0A1B6GMZ3 A0A1B6FJ90 A0A1I8NNP2 A0A1W4VL55 A0A1I8NNP4 A0A1I8NNP3 A0A1I8NNQ1 A0A1I8NNP0 A0A0A9YL57 B4I3D6 B3NYU9 Q32KD3 Q7KSW1 A0A224XGA9 Q494I1 Q9VHW9 A0A0B4KGS8 A0A0B4KGF6 F4WLI9 A0A1I8ND06 A0A0R1E5F7 B4PQX7 A0A0R1E1C0 A0A0R1E1T9 A0A1W4VYN2 A0A1I8ND14 T1PDC2 A0A1I8ND10 A0A1W4VLD5 A0A1W4VXS1 A0A0L0C3D0 A0A1W4VZB2 T1IAQ3 A0A0Q9X183 B3LXB3 A0A0Q9X2Z7 A0A0Q9X880 B4KDE6 I5API7 A0A0Q9X170
A0A067QJA9 A0A2M3YZX2 A0A0P6ISX1 A0A1S4FV30 Q16N63 A0A182K0E0 A0A088AD24 B4R0D3 A0A1B6DSD1 A0A139WP21 D6W798 A0A182LQK5 Q7PEZ3 A0A182NMS5 A0A182HZJ8 A0A182UG76 A0A182GHQ0 A0A182X364 A0A182NZG0 A0A182IL48 U4UQA3 A0A182QPM0 W4VS74 A0A1Y1LQW1 A0A084W110 T1PP11 A0A1W4WFD5 E0VLY1 A0A1B6GN91 A0A0C9Q9W5 A0A0C9RRK4 A0A1B6G0A3 A0A0C9RDI9 A0A232FDY5 A0A1Q3FIR7 A0A1Q3FIE0 B0WFJ4 A0A1J1IIL2 A0A2A3EKW7 K7J8E1 A0A195AUF0 Q1WWE5 A0A3L8DH26 A0A158NN99 E9IVN3 A0A195FWC2 A0A0M8ZUQ0 A0A0J7NJK1 A0A195CT14 E2A542 E2C319 A0A1B0GQM9 A0A026WSX8 A0A151XA78 A0A1B6GMZ3 A0A1B6FJ90 A0A1I8NNP2 A0A1W4VL55 A0A1I8NNP4 A0A1I8NNP3 A0A1I8NNQ1 A0A1I8NNP0 A0A0A9YL57 B4I3D6 B3NYU9 Q32KD3 Q7KSW1 A0A224XGA9 Q494I1 Q9VHW9 A0A0B4KGS8 A0A0B4KGF6 F4WLI9 A0A1I8ND06 A0A0R1E5F7 B4PQX7 A0A0R1E1C0 A0A0R1E1T9 A0A1W4VYN2 A0A1I8ND14 T1PDC2 A0A1I8ND10 A0A1W4VLD5 A0A1W4VXS1 A0A0L0C3D0 A0A1W4VZB2 T1IAQ3 A0A0Q9X183 B3LXB3 A0A0Q9X2Z7 A0A0Q9X880 B4KDE6 I5API7 A0A0Q9X170
Pubmed
19121390
26354079
28756777
24845553
26999592
17510324
+ More
17994087 18362917 19820115 20966253 12364791 14747013 17210077 26483478 23537049 28004739 24438588 20566863 28648823 20075255 30249741 21347285 21282665 20798317 24508170 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 25315136 17550304 26108605 15632085
17994087 18362917 19820115 20966253 12364791 14747013 17210077 26483478 23537049 28004739 24438588 20566863 28648823 20075255 30249741 21347285 21282665 20798317 24508170 25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 25315136 17550304 26108605 15632085
EMBL
BABH01011450
BABH01011451
BABH01011452
BABH01011453
BABH01011454
BABH01011455
+ More
BABH01011456 BABH01011457 NWSH01000329 PCG77366.1 ODYU01009759 SOQ54409.1 RSAL01000013 RVE53393.1 KQ460075 KPJ17935.1 KZ149914 PZC77993.1 KK853280 KDR09039.1 GGFM01001040 MBW21791.1 GDUN01001041 JAN94878.1 CH477837 EAT35779.1 CM000364 EDX12058.1 GEDC01008749 JAS28549.1 KQ971307 KYB29694.1 EFA11040.2 AAAB01008799 EAA45525.5 APCN01001981 JXUM01064328 JXUM01064329 JXUM01064330 JXUM01064331 KQ562293 KXJ76215.1 KB632399 ERL94698.1 AXCN02001208 GANO01000182 JAB59689.1 GEZM01051807 JAV74690.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1 KA650496 AFP65125.1 DS235283 EEB14387.1 GECZ01005892 JAS63877.1 GBYB01011178 JAG80945.1 GBYB01011180 JAG80947.1 GECZ01013907 JAS55862.1 GBYB01011182 JAG80949.1 NNAY01000392 OXU28698.1 GFDL01007672 JAV27373.1 GFDL01007684 JAV27361.1 DS231918 EDS26262.1 CVRI01000047 CRK98297.1 KZ288220 PBC32144.1 KQ976738 KYM75863.1 BT024961 ABE01191.1 QOIP01000008 RLU19482.1 ADTU01021187 GL766328 EFZ15352.1 KQ981208 KYN44743.1 KQ435878 KOX69993.1 LBMM01004233 KMQ92675.1 KQ977372 KYN03269.1 GL436803 EFN71448.1 GL452263 EFN77661.1 AJVK01007411 AJVK01007412 KK107109 EZA59083.1 KQ982351 KYQ57276.1 GECZ01006076 JAS63693.1 GECZ01019503 JAS50266.1 GBHO01009807 JAG33797.1 CH480821 EDW55300.1 CH954181 EDV48212.1 BT023946 AE014297 ABB36450.1 ACZ94845.1 AAF54180.1 GFTR01008936 JAW07490.1 BT023795 AAZ41804.1 ACZ94844.1 AAG22133.2 AGB95745.1 AGB95746.1 GL888208 EGI64925.1 CM000160 KRK03057.1 EDW96301.1 KRK03056.1 KRK03055.1 KA646787 AFP61416.1 JRES01000953 KNC26845.1 ACPB03005729 CH933806 KRG01836.1 CH902617 EDV42757.2 KRG01833.1 KRG01834.1 EDW15955.1 CM000070 EIM52872.2 KRG01835.1
BABH01011456 BABH01011457 NWSH01000329 PCG77366.1 ODYU01009759 SOQ54409.1 RSAL01000013 RVE53393.1 KQ460075 KPJ17935.1 KZ149914 PZC77993.1 KK853280 KDR09039.1 GGFM01001040 MBW21791.1 GDUN01001041 JAN94878.1 CH477837 EAT35779.1 CM000364 EDX12058.1 GEDC01008749 JAS28549.1 KQ971307 KYB29694.1 EFA11040.2 AAAB01008799 EAA45525.5 APCN01001981 JXUM01064328 JXUM01064329 JXUM01064330 JXUM01064331 KQ562293 KXJ76215.1 KB632399 ERL94698.1 AXCN02001208 GANO01000182 JAB59689.1 GEZM01051807 JAV74690.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1 KA650496 AFP65125.1 DS235283 EEB14387.1 GECZ01005892 JAS63877.1 GBYB01011178 JAG80945.1 GBYB01011180 JAG80947.1 GECZ01013907 JAS55862.1 GBYB01011182 JAG80949.1 NNAY01000392 OXU28698.1 GFDL01007672 JAV27373.1 GFDL01007684 JAV27361.1 DS231918 EDS26262.1 CVRI01000047 CRK98297.1 KZ288220 PBC32144.1 KQ976738 KYM75863.1 BT024961 ABE01191.1 QOIP01000008 RLU19482.1 ADTU01021187 GL766328 EFZ15352.1 KQ981208 KYN44743.1 KQ435878 KOX69993.1 LBMM01004233 KMQ92675.1 KQ977372 KYN03269.1 GL436803 EFN71448.1 GL452263 EFN77661.1 AJVK01007411 AJVK01007412 KK107109 EZA59083.1 KQ982351 KYQ57276.1 GECZ01006076 JAS63693.1 GECZ01019503 JAS50266.1 GBHO01009807 JAG33797.1 CH480821 EDW55300.1 CH954181 EDV48212.1 BT023946 AE014297 ABB36450.1 ACZ94845.1 AAF54180.1 GFTR01008936 JAW07490.1 BT023795 AAZ41804.1 ACZ94844.1 AAG22133.2 AGB95745.1 AGB95746.1 GL888208 EGI64925.1 CM000160 KRK03057.1 EDW96301.1 KRK03056.1 KRK03055.1 KA646787 AFP61416.1 JRES01000953 KNC26845.1 ACPB03005729 CH933806 KRG01836.1 CH902617 EDV42757.2 KRG01833.1 KRG01834.1 EDW15955.1 CM000070 EIM52872.2 KRG01835.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000027135
UP000008820
+ More
UP000075881 UP000005203 UP000000304 UP000007266 UP000075882 UP000007062 UP000075884 UP000075840 UP000075902 UP000069940 UP000249989 UP000076407 UP000075885 UP000075880 UP000030742 UP000075886 UP000030765 UP000192223 UP000009046 UP000215335 UP000002320 UP000183832 UP000242457 UP000002358 UP000078540 UP000279307 UP000005205 UP000078541 UP000053105 UP000036403 UP000078542 UP000000311 UP000008237 UP000092462 UP000053097 UP000075809 UP000095300 UP000192221 UP000001292 UP000008711 UP000000803 UP000007755 UP000095301 UP000002282 UP000037069 UP000015103 UP000009192 UP000007801 UP000001819
UP000075881 UP000005203 UP000000304 UP000007266 UP000075882 UP000007062 UP000075884 UP000075840 UP000075902 UP000069940 UP000249989 UP000076407 UP000075885 UP000075880 UP000030742 UP000075886 UP000030765 UP000192223 UP000009046 UP000215335 UP000002320 UP000183832 UP000242457 UP000002358 UP000078540 UP000279307 UP000005205 UP000078541 UP000053105 UP000036403 UP000078542 UP000000311 UP000008237 UP000092462 UP000053097 UP000075809 UP000095300 UP000192221 UP000001292 UP000008711 UP000000803 UP000007755 UP000095301 UP000002282 UP000037069 UP000015103 UP000009192 UP000007801 UP000001819
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JKW4
A0A2A4K015
A0A2H1WMW5
A0A3S2NZW0
A0A194RKS7
A0A2W1C1K1
+ More
A0A067QJA9 A0A2M3YZX2 A0A0P6ISX1 A0A1S4FV30 Q16N63 A0A182K0E0 A0A088AD24 B4R0D3 A0A1B6DSD1 A0A139WP21 D6W798 A0A182LQK5 Q7PEZ3 A0A182NMS5 A0A182HZJ8 A0A182UG76 A0A182GHQ0 A0A182X364 A0A182NZG0 A0A182IL48 U4UQA3 A0A182QPM0 W4VS74 A0A1Y1LQW1 A0A084W110 T1PP11 A0A1W4WFD5 E0VLY1 A0A1B6GN91 A0A0C9Q9W5 A0A0C9RRK4 A0A1B6G0A3 A0A0C9RDI9 A0A232FDY5 A0A1Q3FIR7 A0A1Q3FIE0 B0WFJ4 A0A1J1IIL2 A0A2A3EKW7 K7J8E1 A0A195AUF0 Q1WWE5 A0A3L8DH26 A0A158NN99 E9IVN3 A0A195FWC2 A0A0M8ZUQ0 A0A0J7NJK1 A0A195CT14 E2A542 E2C319 A0A1B0GQM9 A0A026WSX8 A0A151XA78 A0A1B6GMZ3 A0A1B6FJ90 A0A1I8NNP2 A0A1W4VL55 A0A1I8NNP4 A0A1I8NNP3 A0A1I8NNQ1 A0A1I8NNP0 A0A0A9YL57 B4I3D6 B3NYU9 Q32KD3 Q7KSW1 A0A224XGA9 Q494I1 Q9VHW9 A0A0B4KGS8 A0A0B4KGF6 F4WLI9 A0A1I8ND06 A0A0R1E5F7 B4PQX7 A0A0R1E1C0 A0A0R1E1T9 A0A1W4VYN2 A0A1I8ND14 T1PDC2 A0A1I8ND10 A0A1W4VLD5 A0A1W4VXS1 A0A0L0C3D0 A0A1W4VZB2 T1IAQ3 A0A0Q9X183 B3LXB3 A0A0Q9X2Z7 A0A0Q9X880 B4KDE6 I5API7 A0A0Q9X170
A0A067QJA9 A0A2M3YZX2 A0A0P6ISX1 A0A1S4FV30 Q16N63 A0A182K0E0 A0A088AD24 B4R0D3 A0A1B6DSD1 A0A139WP21 D6W798 A0A182LQK5 Q7PEZ3 A0A182NMS5 A0A182HZJ8 A0A182UG76 A0A182GHQ0 A0A182X364 A0A182NZG0 A0A182IL48 U4UQA3 A0A182QPM0 W4VS74 A0A1Y1LQW1 A0A084W110 T1PP11 A0A1W4WFD5 E0VLY1 A0A1B6GN91 A0A0C9Q9W5 A0A0C9RRK4 A0A1B6G0A3 A0A0C9RDI9 A0A232FDY5 A0A1Q3FIR7 A0A1Q3FIE0 B0WFJ4 A0A1J1IIL2 A0A2A3EKW7 K7J8E1 A0A195AUF0 Q1WWE5 A0A3L8DH26 A0A158NN99 E9IVN3 A0A195FWC2 A0A0M8ZUQ0 A0A0J7NJK1 A0A195CT14 E2A542 E2C319 A0A1B0GQM9 A0A026WSX8 A0A151XA78 A0A1B6GMZ3 A0A1B6FJ90 A0A1I8NNP2 A0A1W4VL55 A0A1I8NNP4 A0A1I8NNP3 A0A1I8NNQ1 A0A1I8NNP0 A0A0A9YL57 B4I3D6 B3NYU9 Q32KD3 Q7KSW1 A0A224XGA9 Q494I1 Q9VHW9 A0A0B4KGS8 A0A0B4KGF6 F4WLI9 A0A1I8ND06 A0A0R1E5F7 B4PQX7 A0A0R1E1C0 A0A0R1E1T9 A0A1W4VYN2 A0A1I8ND14 T1PDC2 A0A1I8ND10 A0A1W4VLD5 A0A1W4VXS1 A0A0L0C3D0 A0A1W4VZB2 T1IAQ3 A0A0Q9X183 B3LXB3 A0A0Q9X2Z7 A0A0Q9X880 B4KDE6 I5API7 A0A0Q9X170
Ontologies
GO
PANTHER
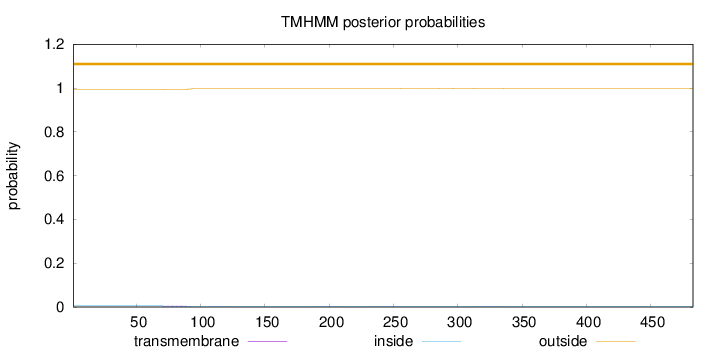
Topology
Length:
483
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12598
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00682
outside
1 - 483
Population Genetic Test Statistics
Pi
278.411096
Theta
172.404781
Tajima's D
1.84398
CLR
0.004609
CSRT
0.852707364631768
Interpretation
Uncertain
