Pre Gene Modal
BGIBMGA010165
Annotation
PREDICTED:_HEAT_repeat-containing_protein_5B_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.004
Sequence
CDS
ATGGCCAACGGAGTACTAACGTCACCTTCAGTATGGAGAGACAATAAAAAAATGAAGTCCGCTTCCCGGGGGCGAGCGCCGGGGAAGCAAATTTATATTAAATGTCTTTCTATTATATTATATTTCATACTAGGTTATTCGGCGGCACTGGCAGCCATACTAGGCGCAGTGCGGCTGTCCCCCCTGGGCGTGCCGCACGGTCGCGGCAAAATCGCCTTTAATGCCGCCGAGCAGTTGCTGCGTTCGGCTGGACAGAGCAGCAGGCTGACGGCTGCCAGAACTAATGCTGGCTGGCTTATTGTCGGCGCCATTTGTACATTAGGGGTCCCGGTGGTGCGAGGTCTCCTACCCCGAATGTTGCTGTTATGGCGTAACAGCTTCCCGAGGTCCGCTAAGGAATTGGAATCGGAAAAATCAAGAGGAGACGCGTTCACTTGGCAGGTGACATTGGAGGGTCGTGCCGGTGCGTTGTCGGCGCTACACAGTCTGTTGATCCACTGTCCGTCGTTGGTCAACAGCGACGACACGGCCAAACGTTTGACGCAGCCCATCGACGGTGCTATCGCTGTGCTCACCAATGTGAACAGCGTAGTCCGCAGTTACGGCGGCGCACTGAAAGCCCCGGCGGCGTTGTTGCGGTTGCGACTGTACCAAACGTGCGCGGCGCTGGGCGGCGCCGGGCCCTCCGCCGCGCCGCTACTACGACTCCTCGCCGCCGAGGTCGCCGGCGCCGCTGACCCGAGCGGGGCTAATGTCGCTACCGGTATGCTAAGGAATGCCATGCATCCACGTGACGTCATACTTCTAGGCGAAGAGGGTTGGATTTATGAAACCGATCATGCAGAAATTGAGGAACAACTCGTATCACCGCTGAGTGCAAGTGGATCAGGAGCATTAGAACACGATCCGTGTTGTCTGTACCGCGGCACTACGGGTGCGCAACCACTGGGCGTCGCGGTTATAGACGCGTCCGTCGTATTGTTCCCGCAAGTATTCGCAAGAGCCGCTAATAAACACAGACAACAAATGCTAGAACACTTCGCGGAATGCATTAAGATGTCTAAGGGTGGTCGCCAGGAAGCAATACAAGTGAATGTGTATACAGCTCTATTATTGGCTCTACGTACTTTAGCAGAGACTAAGACTTCTTTAGGACAAGATCCCGTGAAGAATATTGCCACAGAGTTGATTATTAACGGTCTATCGGCGAACAGCGCCTCAGTACGAGCAGCTGCGGCATCGTGTGCCGGTCGACTCTGCGGCTGCATCACTGAAGCGGAAAGCCAAGCTCTGAGCGAGAAAGTGATAGCATTAGCACGTACAGCCGCCAATCGATCAGTGCGGGCGGCTGCTGCAGCCGCTGCCGGCGCTGTACTCCGGGCTCGAGGCACCGCTTCAAGTGCTACAGCGCTGCCTGTGCTGAGAGCACTCGCACAGGATGCATCTTCTGTCGAGCTTCAGGTGTGGTCAGTACACGCATTGTCGGTGCTGGCGGACGCTTCGGGCCCGATGTTCCGAGCGCACGTCGAGTGGACGCTAGCGCTGGCGCTGAGGCTGATATTCGCCGCTCCGCCTTACCATCACCTGCTCCTGCGCACCGTCGCCAGGCTGCTCGCCGCGCTCATCACTGTGCTCGGACCCGAGTTACAAGTGGTAGCCGTGAGTCGATTCGTTTGCGCGTGCTCGGCGCTGCTGGAGGGCGGCGGCGGCGGCGCGCGCGCTGAGGCCATCGGCTGTCTCCAGCAGCTGCAGATGTTCGCGCCGGCGCACATCAACCTGCACACTCTCGTGCCCAAACTCTGCCACGACTTATCGAACCCGGAGCTGACAGTACGTCGGGCGGCGCTCTGCTGCCTCCGACAACTGTCTCAGAAGGAAGCTTTAGAAGTTTGTCGGTACGCGTTGCTCGCGAAAGATCACGTACCCGCCAAACCGTACTGTGGTGTAGTGATAACGGACACCGGTCTGCCGGGAGCCCTGTTCGCGTTCCTGGACATCGAGCGAGACGAGACCGCGCTGTCGTACGCTAAAGACACACTGACCTGCTGCCTGTTGGCGGCTGCTTCCTCGAACACGATCAGAGACTGGATCGTTCTCGCCAAACGAGTACTCACTGTACGGCTGGAAGACAGCAACAACGCTGACACGGACTTCGACGCGGAAGGCGACGACGACCAGGCGGAGTTCCACGCGGACACGGATCAGTCGACACACCCGGCCGTGCAGTCCCGGTGGCCGACGCGTGTTTTTGCAATGGAGTGCATACAGAAGATAATGGGCGCTTGTGAATTAACTGGCGACAACGCACATTTTGATCTTGTCAAGGCAAAGGAGAAACAAATCGCGGAACCTAATAGTGACTATTTATCGTTCCATCTGTCGGATTTGGTTCGCATGGCGTTCGTCGGCGCCACCGGTGAGTCTGACGCGCTGCGTCTCTGCGGTCTGCGCACGTTGCAGATGATCATACAGCAGTACGCGCGAGCGCCCGAACCGGACTTCCCCGGACATCTGCTGCTGGAACAGTACCAAGCTCAGGTGGGTGCGGCGGTGCGTCCCGCGTTCGCGGGCGACACGGCGTCGCACGTGACGGCGGCCGCGTGCGACGTGTGCTCCGCCTGGATCGGCTGCGGGGTCGCCAGGGACATCAACGACCTCAGGCGAGTTCACCAGCTGCTCGTCTCAAGTCTGGACAAACTGAACACTAAAGGCAACACCACACTCATCTACAACGAGAGTATGGCCACATTGGAAAAGTTATCCATATTAAAGGCCTGGGCTGAGGTATACATCGTGGCGATGGTGAGCAACAACAGCGCTCCGGGCAGCTACGTGAAGCAACTCGACAGCAGACCCTCCAACAATGGAGCTGAACTGGCCAAATGGCGCAACCAAGTACTCCGGGACAACAACGAGATTGACAACGCCGGTCCCGGTAGCGCCGAAGACGACGAGTACGGCGAATTCGAATCTAAAGGCGAGAGTCTACTGAGGCTGGTCGAACCGCAACTGGAGAGCTTGGGCGAGAATTGGATCGCGGCTTTAAAGGATCATGCGCTACTCAGTCTGCCGTCTGGTGAGTTTATGGATAGCTTACTGCTGGTAGGACGGCTTGTGAGTCCACACGAGTAA
Protein
MANGVLTSPSVWRDNKKMKSASRGRAPGKQIYIKCLSIILYFILGYSAALAAILGAVRLSPLGVPHGRGKIAFNAAEQLLRSAGQSSRLTAARTNAGWLIVGAICTLGVPVVRGLLPRMLLLWRNSFPRSAKELESEKSRGDAFTWQVTLEGRAGALSALHSLLIHCPSLVNSDDTAKRLTQPIDGAIAVLTNVNSVVRSYGGALKAPAALLRLRLYQTCAALGGAGPSAAPLLRLLAAEVAGAADPSGANVATGMLRNAMHPRDVILLGEEGWIYETDHAEIEEQLVSPLSASGSGALEHDPCCLYRGTTGAQPLGVAVIDASVVLFPQVFARAANKHRQQMLEHFAECIKMSKGGRQEAIQVNVYTALLLALRTLAETKTSLGQDPVKNIATELIINGLSANSASVRAAAASCAGRLCGCITEAESQALSEKVIALARTAANRSVRAAAAAAAGAVLRARGTASSATALPVLRALAQDASSVELQVWSVHALSVLADASGPMFRAHVEWTLALALRLIFAAPPYHHLLLRTVARLLAALITVLGPELQVVAVSRFVCACSALLEGGGGGARAEAIGCLQQLQMFAPAHINLHTLVPKLCHDLSNPELTVRRAALCCLRQLSQKEALEVCRYALLAKDHVPAKPYCGVVITDTGLPGALFAFLDIERDETALSYAKDTLTCCLLAAASSNTIRDWIVLAKRVLTVRLEDSNNADTDFDAEGDDDQAEFHADTDQSTHPAVQSRWPTRVFAMECIQKIMGACELTGDNAHFDLVKAKEKQIAEPNSDYLSFHLSDLVRMAFVGATGESDALRLCGLRTLQMIIQQYARAPEPDFPGHLLLEQYQAQVGAAVRPAFAGDTASHVTAAACDVCSAWIGCGVARDINDLRRVHQLLVSSLDKLNTKGNTTLIYNESMATLEKLSILKAWAEVYIVAMVSNNSAPGSYVKQLDSRPSNNGAELAKWRNQVLRDNNEIDNAGPGSAEDDEYGEFESKGESLLRLVEPQLESLGENWIAALKDHALLSLPSGEFMDSLLLVGRLVSPHE
Summary
Uniprot
H9JKW4
A0A2H1WMW5
A0A194PZJ2
A0A3S2NZW0
A0A2A4K015
A0A194RKS7
+ More
A0A1Q3FIR7 A0A0P6ISX1 A0A2W1C1K1 A0A182GHQ0 A0A1Y1LQW1 A0A1W4WFD5 A0A1Q3FIE0 A0A1S4FV30 Q16N63 B0WFJ4 W4VS74 A0A067QJA9 D6W798 A0A139WP21 A0A084W110 A0A232FDY5 A0A2A3EKW7 A0A0L7QWS3 K7J8E1 A0A0C9RRK4 A0A0C9Q9W5 A0A0C9RDI9 E2C319 E2A542 A0A154PG50 A0A151IT80 F4WLI9 A0A195CT14 A0A0M8ZUQ0 A0A195FWC2 E9IVN3 A0A158NN99 A0A195AUF0 A0A151XA78 A0A0J7NJK1 A0A1B6GMZ3 A0A1B6LZB4 A0A1B0FR06 A0A1B6MRA5 A0A336KY42 A0A1B6FJ90 A0A1J1IIL2 A0A026WSX8 A0A3L8DH26 E0VLY1 A0A0R3NR15 A0A0R3NJ57 A0A3B0JMY0 B4GDV3 A0A2P8Y6P5 Q295H7 I5API7 A0A0R3NJ45 A0A0R3NJ98 A0A182LQK5 A0A224XGA9 T1IAQ3 T1IQK7 A0A146LS57 A0A0A9YNJ8 A0A0T6B268 A0A0P4W494 A0A1S3H0J5 A0A1S3H0R3 A0A1S3H215 A0A1S3H069 J9JYJ0 R7TVV8 A0A2R5LHZ4 A0A2T7NYT1 A0A093T2H7 A0A091QLB5 A0A091I0K6 A0A1B0GJ56 A0A1S3W8B1 A0A2C9JJA6 A0A091STV1 E1C8J1 A0A087VMI4 A0A091KH53 G1NFC9 A0A093FE98 A0A087RAU4 A0A2I0USX6 A0A091G1H6 A0A0A0A965 A0A091HBJ8 A0A091VQ16 A0A093GAR4 A0A091SIL8 A0A2C9JJB1 A0A093PPU1 A0A091N534 U3KBI2
A0A1Q3FIR7 A0A0P6ISX1 A0A2W1C1K1 A0A182GHQ0 A0A1Y1LQW1 A0A1W4WFD5 A0A1Q3FIE0 A0A1S4FV30 Q16N63 B0WFJ4 W4VS74 A0A067QJA9 D6W798 A0A139WP21 A0A084W110 A0A232FDY5 A0A2A3EKW7 A0A0L7QWS3 K7J8E1 A0A0C9RRK4 A0A0C9Q9W5 A0A0C9RDI9 E2C319 E2A542 A0A154PG50 A0A151IT80 F4WLI9 A0A195CT14 A0A0M8ZUQ0 A0A195FWC2 E9IVN3 A0A158NN99 A0A195AUF0 A0A151XA78 A0A0J7NJK1 A0A1B6GMZ3 A0A1B6LZB4 A0A1B0FR06 A0A1B6MRA5 A0A336KY42 A0A1B6FJ90 A0A1J1IIL2 A0A026WSX8 A0A3L8DH26 E0VLY1 A0A0R3NR15 A0A0R3NJ57 A0A3B0JMY0 B4GDV3 A0A2P8Y6P5 Q295H7 I5API7 A0A0R3NJ45 A0A0R3NJ98 A0A182LQK5 A0A224XGA9 T1IAQ3 T1IQK7 A0A146LS57 A0A0A9YNJ8 A0A0T6B268 A0A0P4W494 A0A1S3H0J5 A0A1S3H0R3 A0A1S3H215 A0A1S3H069 J9JYJ0 R7TVV8 A0A2R5LHZ4 A0A2T7NYT1 A0A093T2H7 A0A091QLB5 A0A091I0K6 A0A1B0GJ56 A0A1S3W8B1 A0A2C9JJA6 A0A091STV1 E1C8J1 A0A087VMI4 A0A091KH53 G1NFC9 A0A093FE98 A0A087RAU4 A0A2I0USX6 A0A091G1H6 A0A0A0A965 A0A091HBJ8 A0A091VQ16 A0A093GAR4 A0A091SIL8 A0A2C9JJB1 A0A093PPU1 A0A091N534 U3KBI2
Pubmed
EMBL
BABH01011450
BABH01011451
BABH01011452
BABH01011453
BABH01011454
BABH01011455
+ More
BABH01011456 BABH01011457 ODYU01009759 SOQ54409.1 KQ459582 KPI98741.1 RSAL01000013 RVE53393.1 NWSH01000329 PCG77366.1 KQ460075 KPJ17935.1 GFDL01007672 JAV27373.1 GDUN01001041 JAN94878.1 KZ149914 PZC77993.1 JXUM01064328 JXUM01064329 JXUM01064330 JXUM01064331 KQ562293 KXJ76215.1 GEZM01051807 JAV74690.1 GFDL01007684 JAV27361.1 CH477837 EAT35779.1 DS231918 EDS26262.1 GANO01000182 JAB59689.1 KK853280 KDR09039.1 KQ971307 EFA11040.2 KYB29694.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1 NNAY01000392 OXU28698.1 KZ288220 PBC32144.1 KQ414706 KOC63073.1 GBYB01011180 JAG80947.1 GBYB01011178 JAG80945.1 GBYB01011182 JAG80949.1 GL452263 EFN77661.1 GL436803 EFN71448.1 KQ434889 KZC10298.1 KQ981037 KYN10152.1 GL888208 EGI64925.1 KQ977372 KYN03269.1 KQ435878 KOX69993.1 KQ981208 KYN44743.1 GL766328 EFZ15352.1 ADTU01021187 KQ976738 KYM75863.1 KQ982351 KYQ57276.1 LBMM01004233 KMQ92675.1 GECZ01006076 JAS63693.1 GEBQ01011003 JAT28974.1 CCAG010006466 GEBQ01001516 JAT38461.1 UFQS01000656 UFQT01000656 SSX05850.1 SSX26209.1 GECZ01019503 JAS50266.1 CVRI01000047 CRK98297.1 KK107109 EZA59083.1 QOIP01000008 RLU19482.1 DS235283 EEB14387.1 CM000070 KRT00998.1 KRT01001.1 OUUW01000007 SPP83584.1 CH479182 EDW34614.1 PYGN01000863 PSN39929.1 EAL28735.2 EIM52872.2 KRT01000.1 KRT00999.1 GFTR01008936 JAW07490.1 ACPB03005729 JH431312 GDHC01009017 JAQ09612.1 GBHO01009805 JAG33799.1 LJIG01016239 KRT81198.1 GDRN01089211 JAI60706.1 ABLF02021124 ABLF02021125 AMQN01010629 KB308278 ELT98043.1 GGLE01005024 MBY09150.1 PZQS01000008 PVD26321.1 KL761273 KFW88924.1 KK799868 KFQ27983.1 KL218009 KFP00960.1 AJWK01019317 KK934195 KFQ46422.1 AADN05000005 KL498729 KFO13826.1 KK743365 KFP39432.1 KK388034 KFV52564.1 KL226263 KFM10598.1 KZ505642 PKU49128.1 KL447561 KFO75009.1 KL870824 KGL89485.1 KL528619 KFO92045.1 KL411249 KFR04785.1 KL215232 KFV63849.1 KK474175 KFQ58467.1 KL415050 KFW78858.1 KK842253 KFP84526.1 AGTO01000393
BABH01011456 BABH01011457 ODYU01009759 SOQ54409.1 KQ459582 KPI98741.1 RSAL01000013 RVE53393.1 NWSH01000329 PCG77366.1 KQ460075 KPJ17935.1 GFDL01007672 JAV27373.1 GDUN01001041 JAN94878.1 KZ149914 PZC77993.1 JXUM01064328 JXUM01064329 JXUM01064330 JXUM01064331 KQ562293 KXJ76215.1 GEZM01051807 JAV74690.1 GFDL01007684 JAV27361.1 CH477837 EAT35779.1 DS231918 EDS26262.1 GANO01000182 JAB59689.1 KK853280 KDR09039.1 KQ971307 EFA11040.2 KYB29694.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1 NNAY01000392 OXU28698.1 KZ288220 PBC32144.1 KQ414706 KOC63073.1 GBYB01011180 JAG80947.1 GBYB01011178 JAG80945.1 GBYB01011182 JAG80949.1 GL452263 EFN77661.1 GL436803 EFN71448.1 KQ434889 KZC10298.1 KQ981037 KYN10152.1 GL888208 EGI64925.1 KQ977372 KYN03269.1 KQ435878 KOX69993.1 KQ981208 KYN44743.1 GL766328 EFZ15352.1 ADTU01021187 KQ976738 KYM75863.1 KQ982351 KYQ57276.1 LBMM01004233 KMQ92675.1 GECZ01006076 JAS63693.1 GEBQ01011003 JAT28974.1 CCAG010006466 GEBQ01001516 JAT38461.1 UFQS01000656 UFQT01000656 SSX05850.1 SSX26209.1 GECZ01019503 JAS50266.1 CVRI01000047 CRK98297.1 KK107109 EZA59083.1 QOIP01000008 RLU19482.1 DS235283 EEB14387.1 CM000070 KRT00998.1 KRT01001.1 OUUW01000007 SPP83584.1 CH479182 EDW34614.1 PYGN01000863 PSN39929.1 EAL28735.2 EIM52872.2 KRT01000.1 KRT00999.1 GFTR01008936 JAW07490.1 ACPB03005729 JH431312 GDHC01009017 JAQ09612.1 GBHO01009805 JAG33799.1 LJIG01016239 KRT81198.1 GDRN01089211 JAI60706.1 ABLF02021124 ABLF02021125 AMQN01010629 KB308278 ELT98043.1 GGLE01005024 MBY09150.1 PZQS01000008 PVD26321.1 KL761273 KFW88924.1 KK799868 KFQ27983.1 KL218009 KFP00960.1 AJWK01019317 KK934195 KFQ46422.1 AADN05000005 KL498729 KFO13826.1 KK743365 KFP39432.1 KK388034 KFV52564.1 KL226263 KFM10598.1 KZ505642 PKU49128.1 KL447561 KFO75009.1 KL870824 KGL89485.1 KL528619 KFO92045.1 KL411249 KFR04785.1 KL215232 KFV63849.1 KK474175 KFQ58467.1 KL415050 KFW78858.1 KK842253 KFP84526.1 AGTO01000393
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000053240
UP000069940
+ More
UP000249989 UP000192223 UP000008820 UP000002320 UP000027135 UP000007266 UP000030765 UP000215335 UP000242457 UP000053825 UP000002358 UP000008237 UP000000311 UP000076502 UP000078492 UP000007755 UP000078542 UP000053105 UP000078541 UP000005205 UP000078540 UP000075809 UP000036403 UP000092444 UP000183832 UP000053097 UP000279307 UP000009046 UP000001819 UP000268350 UP000008744 UP000245037 UP000075882 UP000015103 UP000085678 UP000007819 UP000014760 UP000245119 UP000053258 UP000054308 UP000092461 UP000079721 UP000076420 UP000000539 UP000001645 UP000053286 UP000053760 UP000053858 UP000053283 UP000053875 UP000016665
UP000249989 UP000192223 UP000008820 UP000002320 UP000027135 UP000007266 UP000030765 UP000215335 UP000242457 UP000053825 UP000002358 UP000008237 UP000000311 UP000076502 UP000078492 UP000007755 UP000078542 UP000053105 UP000078541 UP000005205 UP000078540 UP000075809 UP000036403 UP000092444 UP000183832 UP000053097 UP000279307 UP000009046 UP000001819 UP000268350 UP000008744 UP000245037 UP000075882 UP000015103 UP000085678 UP000007819 UP000014760 UP000245119 UP000053258 UP000054308 UP000092461 UP000079721 UP000076420 UP000000539 UP000001645 UP000053286 UP000053760 UP000053858 UP000053283 UP000053875 UP000016665
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9JKW4
A0A2H1WMW5
A0A194PZJ2
A0A3S2NZW0
A0A2A4K015
A0A194RKS7
+ More
A0A1Q3FIR7 A0A0P6ISX1 A0A2W1C1K1 A0A182GHQ0 A0A1Y1LQW1 A0A1W4WFD5 A0A1Q3FIE0 A0A1S4FV30 Q16N63 B0WFJ4 W4VS74 A0A067QJA9 D6W798 A0A139WP21 A0A084W110 A0A232FDY5 A0A2A3EKW7 A0A0L7QWS3 K7J8E1 A0A0C9RRK4 A0A0C9Q9W5 A0A0C9RDI9 E2C319 E2A542 A0A154PG50 A0A151IT80 F4WLI9 A0A195CT14 A0A0M8ZUQ0 A0A195FWC2 E9IVN3 A0A158NN99 A0A195AUF0 A0A151XA78 A0A0J7NJK1 A0A1B6GMZ3 A0A1B6LZB4 A0A1B0FR06 A0A1B6MRA5 A0A336KY42 A0A1B6FJ90 A0A1J1IIL2 A0A026WSX8 A0A3L8DH26 E0VLY1 A0A0R3NR15 A0A0R3NJ57 A0A3B0JMY0 B4GDV3 A0A2P8Y6P5 Q295H7 I5API7 A0A0R3NJ45 A0A0R3NJ98 A0A182LQK5 A0A224XGA9 T1IAQ3 T1IQK7 A0A146LS57 A0A0A9YNJ8 A0A0T6B268 A0A0P4W494 A0A1S3H0J5 A0A1S3H0R3 A0A1S3H215 A0A1S3H069 J9JYJ0 R7TVV8 A0A2R5LHZ4 A0A2T7NYT1 A0A093T2H7 A0A091QLB5 A0A091I0K6 A0A1B0GJ56 A0A1S3W8B1 A0A2C9JJA6 A0A091STV1 E1C8J1 A0A087VMI4 A0A091KH53 G1NFC9 A0A093FE98 A0A087RAU4 A0A2I0USX6 A0A091G1H6 A0A0A0A965 A0A091HBJ8 A0A091VQ16 A0A093GAR4 A0A091SIL8 A0A2C9JJB1 A0A093PPU1 A0A091N534 U3KBI2
A0A1Q3FIR7 A0A0P6ISX1 A0A2W1C1K1 A0A182GHQ0 A0A1Y1LQW1 A0A1W4WFD5 A0A1Q3FIE0 A0A1S4FV30 Q16N63 B0WFJ4 W4VS74 A0A067QJA9 D6W798 A0A139WP21 A0A084W110 A0A232FDY5 A0A2A3EKW7 A0A0L7QWS3 K7J8E1 A0A0C9RRK4 A0A0C9Q9W5 A0A0C9RDI9 E2C319 E2A542 A0A154PG50 A0A151IT80 F4WLI9 A0A195CT14 A0A0M8ZUQ0 A0A195FWC2 E9IVN3 A0A158NN99 A0A195AUF0 A0A151XA78 A0A0J7NJK1 A0A1B6GMZ3 A0A1B6LZB4 A0A1B0FR06 A0A1B6MRA5 A0A336KY42 A0A1B6FJ90 A0A1J1IIL2 A0A026WSX8 A0A3L8DH26 E0VLY1 A0A0R3NR15 A0A0R3NJ57 A0A3B0JMY0 B4GDV3 A0A2P8Y6P5 Q295H7 I5API7 A0A0R3NJ45 A0A0R3NJ98 A0A182LQK5 A0A224XGA9 T1IAQ3 T1IQK7 A0A146LS57 A0A0A9YNJ8 A0A0T6B268 A0A0P4W494 A0A1S3H0J5 A0A1S3H0R3 A0A1S3H215 A0A1S3H069 J9JYJ0 R7TVV8 A0A2R5LHZ4 A0A2T7NYT1 A0A093T2H7 A0A091QLB5 A0A091I0K6 A0A1B0GJ56 A0A1S3W8B1 A0A2C9JJA6 A0A091STV1 E1C8J1 A0A087VMI4 A0A091KH53 G1NFC9 A0A093FE98 A0A087RAU4 A0A2I0USX6 A0A091G1H6 A0A0A0A965 A0A091HBJ8 A0A091VQ16 A0A093GAR4 A0A091SIL8 A0A2C9JJB1 A0A093PPU1 A0A091N534 U3KBI2
Ontologies
GO
PANTHER
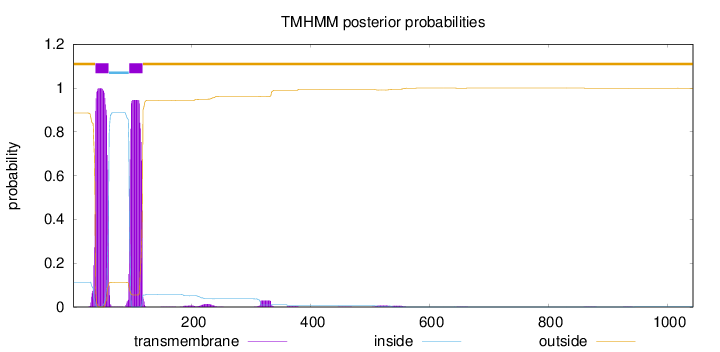
Topology
Length:
1043
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.9795600000001
Exp number, first 60 AAs:
22.38621
Total prob of N-in:
0.11354
POSSIBLE N-term signal
sequence
outside
1 - 37
TMhelix
38 - 60
inside
61 - 94
TMhelix
95 - 117
outside
118 - 1043
Population Genetic Test Statistics
Pi
374.005772
Theta
173.001566
Tajima's D
3.154362
CLR
0.356173
CSRT
0.985100744962752
Interpretation
Uncertain
