Pre Gene Modal
BGIBMGA010165
Annotation
PREDICTED:_HEAT_repeat-containing_protein_5B_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.196 Nuclear Reliability : 1.771
Sequence
CDS
ATGGAGGCGTTATGCGCGCAACGCGACATGAGCGAGGAAAATACGATATCTGTTTTGCTCTCGCTGGTCACCCTACTCGACGCTCCCGACAACAGAGCCAGGCTACTCAGAGACAAAGCATTAGCTATCGAGCTGTGTCAGATAATACATCGCACGGCTCTAACTCAACCAAGTATAGAGGCGTTGTTACTGTCCGCCGACGTTCTGCGTTTGGTGATCGACGCGGCCAGAGAACAATTGCATGCCAACATGCAAGCTAAAATTAAAGAACTGGCACCTAACGAAGCATCAGACGGATCAATCCCTCAATCAGTTCTCGAGGTGGTCCACACGTTAGGCGAAGGAGGTCCTACAGGGGACCTTCCGCCGGGGAAGTCTTTAGTGTTCGCTTGTCTGGAGGTCTGCGTCTGCTTACTTGTGCGGCGCCTGCCCTCCTTGAGTCCGGTGAGGCAGGAGGGCAAGGTCGGGGTCATAGGAGTGCCCAAAGGACTTGGAATACATGGCGATACGTTGTTGGTCAAAAGCTTGGCTGCTATGTCCGAGTTGCTGTCTTTAACTAGTCCCCAGGGCGCGCTCTCAATATTACCGACGATACTGTACCTATCGACCGAGGTGCTGCAAGAGAGTAAAGACTCGCCGACGGTGGCGGACGCGGGCGTGCTGCTGTTGCAGCGCGCGGCCGAGTGCCGGGCGGTGCGCGTGCACGACCACACGCGCGAGCAGCACGACAAGCTGCTGCAGAGCACGCTCGCCAAACTAGTCGAGCGAGTCAAGAGCGATGATAAGGAATGTCGTTTGGACAGCATTACGGGTGCACGCGGTATGGCCGTCGTATTGAATGCTGCCGAACCGATACCCGCTCTCCATTATCCTTGCATCAATCATTTCAGACAATGTCTCGATACCGACGATCACGAGCGGTTCGCGTCGTGTTGCGCGGTGGTGCGCAGCGTGTGGGGCGGGGGCGCGGAGGCGCGGCGCGTGTCATGGTGGGCGCGCGCGCTGGCGGCGCGGCTGGTGGCGCGCGCACACGCCGCCCGCCGCCCCGCGCACGCGCCGCACGCCCGCGCCGCGCTCGCCGCCCTGCACGCGCTCGACGACCTGGTGCGGGCCGCGCCGCCTCAACCGCGAATTCAACTTCTCTGGTTACTGGTGCCGATAGTGATTTCGTATCTACGGGAAGGGAACGAACTGAGCCAAGCCCCGCCCCACGTTCGTACGTTGCACGACTTCGCCCTGCAATGGCTAACGAAGATCGGACCTAAATATCCCCAGGAATTCAAAACGATGATGCAACAGTCTTCGGAGCTGCGCAATAAGTTAGAGAGCGCTCTGAAGTCGAGTCAGAGCGGGAATCGTTCGGGCCGCCCGACGCAACAGCAGAGGCCCGTGTTCGACGCGAAGCCGACTAAACCTACTATACAACTTAAAACGGACTTCAGCGATTTCAGATAA
Protein
MEALCAQRDMSEENTISVLLSLVTLLDAPDNRARLLRDKALAIELCQIIHRTALTQPSIEALLLSADVLRLVIDAAREQLHANMQAKIKELAPNEASDGSIPQSVLEVVHTLGEGGPTGDLPPGKSLVFACLEVCVCLLVRRLPSLSPVRQEGKVGVIGVPKGLGIHGDTLLVKSLAAMSELLSLTSPQGALSILPTILYLSTEVLQESKDSPTVADAGVLLLQRAAECRAVRVHDHTREQHDKLLQSTLAKLVERVKSDDKECRLDSITGARGMAVVLNAAEPIPALHYPCINHFRQCLDTDDHERFASCCAVVRSVWGGGAEARRVSWWARALAARLVARAHAARRPAHAPHARAALAALHALDDLVRAAPPQPRIQLLWLLVPIVISYLREGNELSQAPPHVRTLHDFALQWLTKIGPKYPQEFKTMMQQSSELRNKLESALKSSQSGNRSGRPTQQQRPVFDAKPTKPTIQLKTDFSDFR
Summary
Uniprot
A0A2H1WMW5
A0A3S2NZW0
A0A2A4K015
A0A034VSR0
D6W798
A0A0C9Q9W5
+ More
A0A0C9RDI9 A0A139WP21 A0A0Q9X183 B4KDE6 A0A0K8VWF8 A0A0R1E5F7 B4JUR9 A0A1W4WFD5 B4PQX7 A0A1B0BA82 A0A1A9XW67 B3NYU9 A0A0Q9X880 B4I3D6 A0A0A1XSH2 B4GDV3 A0A034VUH6 A0A0A1WWY5 A0A034VTR1 A0A034VQB8 A0A0Q9X2Z7 A0A0R3NJ57 Q295H7 A0A0Q9X170 A0A1B0A1P3 A0A1W4VZB2 A0A1B0FR06 A0A1W4VYN2 A0A336KY42 Q7KSW1 Q32KD3 A0A1I8ND14 Q494I1 A0A1I8ND10 Q9VHW9 A0A0R1E1T9 A0A1I8ND06 A0A0R1E1C0 T1PDC2 B3LXB3 A0A3B0JMY0 B4M569 A0A1I8NNP3 A0A1I8NNP2 E2C319 A0A0R3NR15 A0A1W4VL55 I5API7 A0A1W4VLD5 A0A0R3NJ45 A0A1W4VXS1 A0A0M3QY91 A0A0L0C3D0 A0A1A9X2M8 A0A0B4KGF6 A0A0B4KGS8 A0A1B6INV9 W4VS74 W8AYN3 W8BDT9 W8B2R2 W8AYN5 A0A034VSR4 A0A034VUI1 A0A1B6FJ90 A0A1B6GMZ3 A0A0P6ISX1 W5JTU9 Q16N63 A0A1S4FV30 A0A1Q3FIR7 A0A1Q3FIE0 A0A2M4A3P2 A0A2M4B848 A0A2M4B851 B0WFJ4 B4NJ40 A0A182QPM0 A0A0Q9WUT0 A0A182Y4M5 A0A2M4A3K1 A0A2M3YZX2 A0A182NZG0 A0A0Q9WV78 A0A182VRX2 A0A0Q9WV26 A0A0T6B268 A0A182IL48 A0A084W110
A0A0C9RDI9 A0A139WP21 A0A0Q9X183 B4KDE6 A0A0K8VWF8 A0A0R1E5F7 B4JUR9 A0A1W4WFD5 B4PQX7 A0A1B0BA82 A0A1A9XW67 B3NYU9 A0A0Q9X880 B4I3D6 A0A0A1XSH2 B4GDV3 A0A034VUH6 A0A0A1WWY5 A0A034VTR1 A0A034VQB8 A0A0Q9X2Z7 A0A0R3NJ57 Q295H7 A0A0Q9X170 A0A1B0A1P3 A0A1W4VZB2 A0A1B0FR06 A0A1W4VYN2 A0A336KY42 Q7KSW1 Q32KD3 A0A1I8ND14 Q494I1 A0A1I8ND10 Q9VHW9 A0A0R1E1T9 A0A1I8ND06 A0A0R1E1C0 T1PDC2 B3LXB3 A0A3B0JMY0 B4M569 A0A1I8NNP3 A0A1I8NNP2 E2C319 A0A0R3NR15 A0A1W4VL55 I5API7 A0A1W4VLD5 A0A0R3NJ45 A0A1W4VXS1 A0A0M3QY91 A0A0L0C3D0 A0A1A9X2M8 A0A0B4KGF6 A0A0B4KGS8 A0A1B6INV9 W4VS74 W8AYN3 W8BDT9 W8B2R2 W8AYN5 A0A034VSR4 A0A034VUI1 A0A1B6FJ90 A0A1B6GMZ3 A0A0P6ISX1 W5JTU9 Q16N63 A0A1S4FV30 A0A1Q3FIR7 A0A1Q3FIE0 A0A2M4A3P2 A0A2M4B848 A0A2M4B851 B0WFJ4 B4NJ40 A0A182QPM0 A0A0Q9WUT0 A0A182Y4M5 A0A2M4A3K1 A0A2M3YZX2 A0A182NZG0 A0A0Q9WV78 A0A182VRX2 A0A0Q9WV26 A0A0T6B268 A0A182IL48 A0A084W110
Pubmed
EMBL
ODYU01009759
SOQ54409.1
RSAL01000013
RVE53393.1
NWSH01000329
PCG77366.1
+ More
GAKP01013453 JAC45499.1 KQ971307 EFA11040.2 GBYB01011178 JAG80945.1 GBYB01011182 JAG80949.1 KYB29694.1 CH933806 KRG01836.1 EDW15955.1 GDHF01009105 JAI43209.1 CM000160 KRK03057.1 CH916374 EDV91239.1 EDW96301.1 JXJN01010867 CH954181 EDV48212.1 KRG01834.1 CH480821 EDW55300.1 GBXI01000043 JAD14249.1 CH479182 EDW34614.1 GAKP01013452 JAC45500.1 GBXI01010965 JAD03327.1 GAKP01013445 JAC45507.1 GAKP01013446 JAC45506.1 KRG01833.1 CM000070 KRT01001.1 EAL28735.2 KRG01835.1 CCAG010006466 UFQS01000656 UFQT01000656 SSX05850.1 SSX26209.1 AE014297 AAF54180.1 BT023946 ABB36450.1 ACZ94845.1 BT023795 AAZ41804.1 ACZ94844.1 AAG22133.2 KRK03055.1 KRK03056.1 KA646787 AFP61416.1 CH902617 EDV42757.2 OUUW01000007 SPP83584.1 CH940652 EDW59780.2 GL452263 EFN77661.1 KRT00998.1 EIM52872.2 KRT01000.1 CP012526 ALC47209.1 JRES01000953 KNC26845.1 AGB95746.1 AGB95745.1 GECU01019091 JAS88615.1 GANO01000182 JAB59689.1 GAMC01015228 GAMC01015224 JAB91327.1 GAMC01015229 JAB91326.1 GAMC01015227 JAB91328.1 GAMC01015223 JAB91332.1 GAKP01013448 JAC45504.1 GAKP01013447 JAC45505.1 GECZ01019503 JAS50266.1 GECZ01006076 JAS63693.1 GDUN01001041 JAN94878.1 ADMH02000156 ETN67556.1 CH477837 EAT35779.1 GFDL01007672 JAV27373.1 GFDL01007684 JAV27361.1 GGFK01001937 MBW35258.1 GGFJ01000083 MBW49224.1 GGFJ01000082 MBW49223.1 DS231918 EDS26262.1 CH964272 EDW83833.1 AXCN02001208 KRF99898.1 GGFK01002062 MBW35383.1 GGFM01001040 MBW21791.1 KRF99900.1 KRF99899.1 LJIG01016239 KRT81198.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1
GAKP01013453 JAC45499.1 KQ971307 EFA11040.2 GBYB01011178 JAG80945.1 GBYB01011182 JAG80949.1 KYB29694.1 CH933806 KRG01836.1 EDW15955.1 GDHF01009105 JAI43209.1 CM000160 KRK03057.1 CH916374 EDV91239.1 EDW96301.1 JXJN01010867 CH954181 EDV48212.1 KRG01834.1 CH480821 EDW55300.1 GBXI01000043 JAD14249.1 CH479182 EDW34614.1 GAKP01013452 JAC45500.1 GBXI01010965 JAD03327.1 GAKP01013445 JAC45507.1 GAKP01013446 JAC45506.1 KRG01833.1 CM000070 KRT01001.1 EAL28735.2 KRG01835.1 CCAG010006466 UFQS01000656 UFQT01000656 SSX05850.1 SSX26209.1 AE014297 AAF54180.1 BT023946 ABB36450.1 ACZ94845.1 BT023795 AAZ41804.1 ACZ94844.1 AAG22133.2 KRK03055.1 KRK03056.1 KA646787 AFP61416.1 CH902617 EDV42757.2 OUUW01000007 SPP83584.1 CH940652 EDW59780.2 GL452263 EFN77661.1 KRT00998.1 EIM52872.2 KRT01000.1 CP012526 ALC47209.1 JRES01000953 KNC26845.1 AGB95746.1 AGB95745.1 GECU01019091 JAS88615.1 GANO01000182 JAB59689.1 GAMC01015228 GAMC01015224 JAB91327.1 GAMC01015229 JAB91326.1 GAMC01015227 JAB91328.1 GAMC01015223 JAB91332.1 GAKP01013448 JAC45504.1 GAKP01013447 JAC45505.1 GECZ01019503 JAS50266.1 GECZ01006076 JAS63693.1 GDUN01001041 JAN94878.1 ADMH02000156 ETN67556.1 CH477837 EAT35779.1 GFDL01007672 JAV27373.1 GFDL01007684 JAV27361.1 GGFK01001937 MBW35258.1 GGFJ01000083 MBW49224.1 GGFJ01000082 MBW49223.1 DS231918 EDS26262.1 CH964272 EDW83833.1 AXCN02001208 KRF99898.1 GGFK01002062 MBW35383.1 GGFM01001040 MBW21791.1 KRF99900.1 KRF99899.1 LJIG01016239 KRT81198.1 ATLV01019181 ATLV01019182 KE525264 KFB43904.1
Proteomes
UP000283053
UP000218220
UP000007266
UP000009192
UP000002282
UP000001070
+ More
UP000192223 UP000092460 UP000092443 UP000008711 UP000001292 UP000008744 UP000001819 UP000092445 UP000192221 UP000092444 UP000000803 UP000095301 UP000007801 UP000268350 UP000008792 UP000095300 UP000008237 UP000092553 UP000037069 UP000091820 UP000000673 UP000008820 UP000002320 UP000007798 UP000075886 UP000076408 UP000075885 UP000075920 UP000075880 UP000030765
UP000192223 UP000092460 UP000092443 UP000008711 UP000001292 UP000008744 UP000001819 UP000092445 UP000192221 UP000092444 UP000000803 UP000095301 UP000007801 UP000268350 UP000008792 UP000095300 UP000008237 UP000092553 UP000037069 UP000091820 UP000000673 UP000008820 UP000002320 UP000007798 UP000075886 UP000076408 UP000075885 UP000075920 UP000075880 UP000030765
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2H1WMW5
A0A3S2NZW0
A0A2A4K015
A0A034VSR0
D6W798
A0A0C9Q9W5
+ More
A0A0C9RDI9 A0A139WP21 A0A0Q9X183 B4KDE6 A0A0K8VWF8 A0A0R1E5F7 B4JUR9 A0A1W4WFD5 B4PQX7 A0A1B0BA82 A0A1A9XW67 B3NYU9 A0A0Q9X880 B4I3D6 A0A0A1XSH2 B4GDV3 A0A034VUH6 A0A0A1WWY5 A0A034VTR1 A0A034VQB8 A0A0Q9X2Z7 A0A0R3NJ57 Q295H7 A0A0Q9X170 A0A1B0A1P3 A0A1W4VZB2 A0A1B0FR06 A0A1W4VYN2 A0A336KY42 Q7KSW1 Q32KD3 A0A1I8ND14 Q494I1 A0A1I8ND10 Q9VHW9 A0A0R1E1T9 A0A1I8ND06 A0A0R1E1C0 T1PDC2 B3LXB3 A0A3B0JMY0 B4M569 A0A1I8NNP3 A0A1I8NNP2 E2C319 A0A0R3NR15 A0A1W4VL55 I5API7 A0A1W4VLD5 A0A0R3NJ45 A0A1W4VXS1 A0A0M3QY91 A0A0L0C3D0 A0A1A9X2M8 A0A0B4KGF6 A0A0B4KGS8 A0A1B6INV9 W4VS74 W8AYN3 W8BDT9 W8B2R2 W8AYN5 A0A034VSR4 A0A034VUI1 A0A1B6FJ90 A0A1B6GMZ3 A0A0P6ISX1 W5JTU9 Q16N63 A0A1S4FV30 A0A1Q3FIR7 A0A1Q3FIE0 A0A2M4A3P2 A0A2M4B848 A0A2M4B851 B0WFJ4 B4NJ40 A0A182QPM0 A0A0Q9WUT0 A0A182Y4M5 A0A2M4A3K1 A0A2M3YZX2 A0A182NZG0 A0A0Q9WV78 A0A182VRX2 A0A0Q9WV26 A0A0T6B268 A0A182IL48 A0A084W110
A0A0C9RDI9 A0A139WP21 A0A0Q9X183 B4KDE6 A0A0K8VWF8 A0A0R1E5F7 B4JUR9 A0A1W4WFD5 B4PQX7 A0A1B0BA82 A0A1A9XW67 B3NYU9 A0A0Q9X880 B4I3D6 A0A0A1XSH2 B4GDV3 A0A034VUH6 A0A0A1WWY5 A0A034VTR1 A0A034VQB8 A0A0Q9X2Z7 A0A0R3NJ57 Q295H7 A0A0Q9X170 A0A1B0A1P3 A0A1W4VZB2 A0A1B0FR06 A0A1W4VYN2 A0A336KY42 Q7KSW1 Q32KD3 A0A1I8ND14 Q494I1 A0A1I8ND10 Q9VHW9 A0A0R1E1T9 A0A1I8ND06 A0A0R1E1C0 T1PDC2 B3LXB3 A0A3B0JMY0 B4M569 A0A1I8NNP3 A0A1I8NNP2 E2C319 A0A0R3NR15 A0A1W4VL55 I5API7 A0A1W4VLD5 A0A0R3NJ45 A0A1W4VXS1 A0A0M3QY91 A0A0L0C3D0 A0A1A9X2M8 A0A0B4KGF6 A0A0B4KGS8 A0A1B6INV9 W4VS74 W8AYN3 W8BDT9 W8B2R2 W8AYN5 A0A034VSR4 A0A034VUI1 A0A1B6FJ90 A0A1B6GMZ3 A0A0P6ISX1 W5JTU9 Q16N63 A0A1S4FV30 A0A1Q3FIR7 A0A1Q3FIE0 A0A2M4A3P2 A0A2M4B848 A0A2M4B851 B0WFJ4 B4NJ40 A0A182QPM0 A0A0Q9WUT0 A0A182Y4M5 A0A2M4A3K1 A0A2M3YZX2 A0A182NZG0 A0A0Q9WV78 A0A182VRX2 A0A0Q9WV26 A0A0T6B268 A0A182IL48 A0A084W110
Ontologies
GO
PANTHER
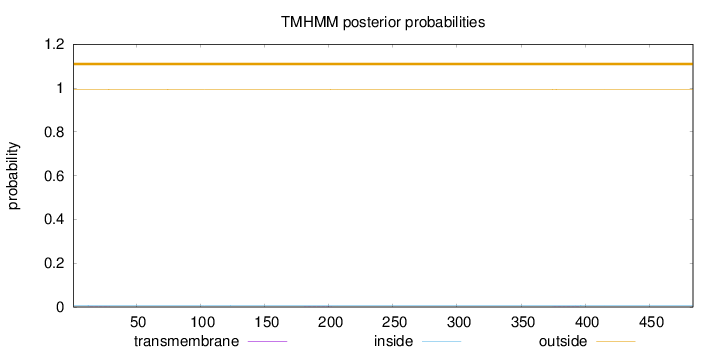
Topology
Length:
484
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02555
Exp number, first 60 AAs:
0.0044
Total prob of N-in:
0.00633
outside
1 - 484
Population Genetic Test Statistics
Pi
300.25086
Theta
174.170943
Tajima's D
2.296058
CLR
0.232129
CSRT
0.924503774811259
Interpretation
Uncertain
