Pre Gene Modal
BGIBMGA010207
Annotation
tRNA_methyltransferase_[Danaus_plexippus]
Full name
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A
Location in the cell
Cytoplasmic Reliability : 2.593
Sequence
CDS
ATGAGTTTTAGAAATTATAAGGATAAAGTAGATGAAGGAGATACAGTGATCTTATACCTTAGTAACAATTTATATGCAATAGATGTTAAGACTGAAGTTAAAAATAAGAAAGGTGAAACGATTGAGAATGTCTTCCAAACACCTTTCGGTGCTTTAAAAGTAAGGAACTTAATTGGTATACAGTATGGAAGTCGGATAGAACTATCAAAGGGCTGGGGACACATACTACAGCCAACCCCTGAGATGTGGTCCCTAACATTGCCTCACCGCACTCAAATTATATACACGCCTGACATAAGCATGATCCTGTTCCAATTGGACTTAGTTCCTGGCTGTGTTGTCATCGAAGCTGGTACAGGTAGTGGCTCCTTAACTCATGCACTCATACGTCGCGTGCGGCCAGAAGGGCATGTTTACACTTTTGATTTTCATGAGCACAGATCCAAAATTGCTAGAGAAGAGTTTGAGGCTCATGATGTTGCCGACTTTGTTACTGCGCAACACAGAGACATCATACGAGACGGCTTTTCTGAAGAATTGAATGGGAAAGCGGATGCCGTGTTTTTGGATTTACCCAGTCCTTGGCTTGGAATTTCTCATGCAGTAAAAGCCATCAAAGAACAAGGCGGAAGATTCTGTTCGTTCTCTCCTTGCATCGAACAAGTACAAAGGACATGTCTAGTTCTTCAAGAGAATGGCTTCCAGGACATCAACACTATGGAGATATTGCAGAGGGAGCTTAAAGTCACTAAAAGAACGATACCCGTGAGGGACTTATCATTTCTGAAACACAAAATCTCTTAG
Protein
MSFRNYKDKVDEGDTVILYLSNNLYAIDVKTEVKNKKGETIENVFQTPFGALKVRNLIGIQYGSRIELSKGWGHILQPTPEMWSLTLPHRTQIIYTPDISMILFQLDLVPGCVVIEAGTGSGSLTHALIRRVRPEGHVYTFDFHEHRSKIAREEFEAHDVADFVTAQHRDIIRDGFSEELNGKADAVFLDLPSPWLGISHAVKAIKEQGGRFCSFSPCIEQVQRTCLVLQENGFQDINTMEILQRELKVTKRTIPVRDLSFLKHKIS
Summary
Description
Catalytic subunit of tRNA (adenine-N(1)-)-methyltransferase, which catalyzes the formation of N(1)-methyladenine at position 58 (m1A58) in initiator methionyl-tRNA.
Catalytic Activity
adenosine(58) in tRNA + S-adenosyl-L-methionine = H(+) + N(1)-methyladenosine(58) in tRNA + S-adenosyl-L-homocysteine
Similarity
Belongs to the class I-like SAM-binding methyltransferase superfamily. TRM61 family.
Uniprot
A0A212ES41
A0A2A4JXF9
A0A2W1BZ81
H9JL06
A0A3S2M7R5
A0A2H1WYW7
+ More
A0A0L0C3C8 W8BQM5 A0A0K8VKE8 R4FND9 A0A0P4VVB4 A0A224XTM2 A0A023F8C2 A0A1I8PCD6 A0A1I8MBH5 A0A1B6FJX9 N6UJJ4 A0A1Q3EXW5 B0WJB3 Q16ZN4 U4UIK8 A0A182H750 A0A2P8Z6H3 D6WA21 A0A1L8DGP9 K7J3Z8 A0A336LJD7 A0A1B6CPE4 A0A1Y1M862 A0A0C9QQW2 A0A1A9WF52 A0A0M3QXZ4 A0A194Q178 A0A026WXM3 A0A1W4V7K6 B3M2U0 B4LYT4 B4NK44 B3P698 Q9VBH7 B4JGK1 B4K6E2 B4QW28 A0A1B3PDB5 A0A0L7R9V6 B4ICJ3 B4PSW4 A0A151X951 A0A182XXM9 E2ALA4 Q296Z9 B4GEF2 A0A1J1HHM2 A0A1A9XEX3 A0A1B0BEV8 A0A195FBW0 Q7Q294 A0A1A9VDZ9 A0A154P0A2 A0A158NKQ3 A0A195DG69 A0A2M4CTN8 A0A195CKY4 A0A182JCP4 V9IIE3 A0A232EVS8 A0A310SNV1 A0A0A9Y1D5 T1DPM6 F4X3G2 A0A088AD89 A0A3B0K611 A0A0M8ZYV7 A0A1B3PDJ1 A0A0P4Y650 A0A2S2PZ83 A0A0P5TII0 E0W403 A0A3L8D9N4 A0A0P6EWY4 A0A1B0GF17 A0A2A3ECL0 A0A2S2NYU5 A0A2H8TFL0 A0A0P6D1T9 A0A182QCC9 E2C811 A0A195BLV4 E9GX16 A0A0P4WSQ9 A0A2J7QT15 A0A1B0DQK6 A0A0P5UAJ4 A0A1S3H9R5 A0A0P5BIN1 T1HIF1 A0A067RDJ5 A0A131XWK8 T1J699 A0A1S3H7W8
A0A0L0C3C8 W8BQM5 A0A0K8VKE8 R4FND9 A0A0P4VVB4 A0A224XTM2 A0A023F8C2 A0A1I8PCD6 A0A1I8MBH5 A0A1B6FJX9 N6UJJ4 A0A1Q3EXW5 B0WJB3 Q16ZN4 U4UIK8 A0A182H750 A0A2P8Z6H3 D6WA21 A0A1L8DGP9 K7J3Z8 A0A336LJD7 A0A1B6CPE4 A0A1Y1M862 A0A0C9QQW2 A0A1A9WF52 A0A0M3QXZ4 A0A194Q178 A0A026WXM3 A0A1W4V7K6 B3M2U0 B4LYT4 B4NK44 B3P698 Q9VBH7 B4JGK1 B4K6E2 B4QW28 A0A1B3PDB5 A0A0L7R9V6 B4ICJ3 B4PSW4 A0A151X951 A0A182XXM9 E2ALA4 Q296Z9 B4GEF2 A0A1J1HHM2 A0A1A9XEX3 A0A1B0BEV8 A0A195FBW0 Q7Q294 A0A1A9VDZ9 A0A154P0A2 A0A158NKQ3 A0A195DG69 A0A2M4CTN8 A0A195CKY4 A0A182JCP4 V9IIE3 A0A232EVS8 A0A310SNV1 A0A0A9Y1D5 T1DPM6 F4X3G2 A0A088AD89 A0A3B0K611 A0A0M8ZYV7 A0A1B3PDJ1 A0A0P4Y650 A0A2S2PZ83 A0A0P5TII0 E0W403 A0A3L8D9N4 A0A0P6EWY4 A0A1B0GF17 A0A2A3ECL0 A0A2S2NYU5 A0A2H8TFL0 A0A0P6D1T9 A0A182QCC9 E2C811 A0A195BLV4 E9GX16 A0A0P4WSQ9 A0A2J7QT15 A0A1B0DQK6 A0A0P5UAJ4 A0A1S3H9R5 A0A0P5BIN1 T1HIF1 A0A067RDJ5 A0A131XWK8 T1J699 A0A1S3H7W8
EC Number
2.1.1.220
Pubmed
22118469
28756777
19121390
26108605
24495485
27129103
+ More
25474469 25315136 23537049 17510324 26483478 29403074 18362917 19820115 20075255 28004739 26354079 24508170 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20798317 15632085 12364791 14747013 17210077 21347285 28648823 25401762 26823975 21719571 20566863 30249741 21292972 24845553
25474469 25315136 23537049 17510324 26483478 29403074 18362917 19820115 20075255 28004739 26354079 24508170 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20798317 15632085 12364791 14747013 17210077 21347285 28648823 25401762 26823975 21719571 20566863 30249741 21292972 24845553
EMBL
AGBW02012891
OWR44271.1
NWSH01000396
PCG76705.1
KZ149914
PZC77996.1
+ More
BABH01011443 RSAL01000013 RVE53392.1 ODYU01012082 SOQ58192.1 JRES01000953 KNC26838.1 GAMC01005193 JAC01363.1 GDHF01012953 JAI39361.1 GAHY01001334 JAA76176.1 GDKW01000610 JAI55985.1 GFTR01004973 JAW11453.1 GBBI01001172 JAC17540.1 GECZ01019288 JAS50481.1 APGK01031799 APGK01031800 KB740824 ENN78837.1 GFDL01014915 JAV20130.1 DS231958 EDS29048.1 CH477487 EAT40124.1 KB632314 ERL92238.1 JXUM01028000 KQ560807 KXJ80807.1 PYGN01000174 PSN52092.1 KQ971312 EEZ98075.1 GFDF01008452 JAV05632.1 AAZX01000987 UFQT01000022 SSX18016.1 GEDC01021988 JAS15310.1 GEZM01041321 GEZM01041320 JAV80515.1 GBYB01006119 JAG75886.1 CP012526 ALC46729.1 KQ459582 KPI98744.1 KK107078 EZA60491.1 CH902617 EDV43470.1 CH940650 EDW67011.1 CH964272 EDW85086.1 CH954182 EDV53568.1 AE014297 AY119248 AAF56565.1 AAM51108.1 ALI30641.1 CH916369 EDV93698.1 CH933806 EDW15215.1 CM000364 EDX14527.1 KU659911 AOG17710.1 KQ414620 KOC67652.1 CH480828 EDW45089.1 CM000160 EDW98651.1 KQ982409 KYQ56818.1 GL440570 EFN65783.1 CM000070 EAL28307.1 CH479182 EDW33987.1 CVRI01000004 CRK87527.1 JXJN01013032 KQ981693 KYN37871.1 AAAB01008978 EAA13617.4 KQ434787 KZC05345.1 ADTU01018960 KQ980886 KYN11837.1 GGFL01004536 MBW68714.1 KQ977649 KYN00739.1 JR045824 AEY60064.1 NNAY01001960 OXU22446.1 KQ760551 OAD59820.1 GBHO01018153 GDHC01016527 GDHC01013376 JAG25451.1 JAQ02102.1 JAQ05253.1 GAMD01003219 JAA98371.1 GL888613 EGI58990.1 OUUW01000023 SPP89565.1 KQ435811 KOX72896.1 KU659988 AOG17786.1 GDIP01235789 JAI87612.1 GGMS01001633 MBY70836.1 GDIP01127358 JAL76356.1 AAZO01007496 DS235885 EEB20359.1 QOIP01000011 RLU17220.1 GDIQ01056923 JAN37814.1 CCAG010012732 KZ288285 PBC29475.1 GGMR01009519 MBY22138.1 GFXV01001046 MBW12851.1 GDIQ01086408 JAN08329.1 AXCN02000286 GL453506 EFN75929.1 KQ976439 KYM86709.1 GL732571 EFX75994.1 GDRN01029702 GDRN01029701 JAI67589.1 NEVH01011202 PNF31726.1 AJVK01019160 GDIP01115761 JAL87953.1 GDIP01188298 JAJ35104.1 ACPB03009370 KK852771 KDR16889.1 GEFM01005188 JAP70608.1 JH431874
BABH01011443 RSAL01000013 RVE53392.1 ODYU01012082 SOQ58192.1 JRES01000953 KNC26838.1 GAMC01005193 JAC01363.1 GDHF01012953 JAI39361.1 GAHY01001334 JAA76176.1 GDKW01000610 JAI55985.1 GFTR01004973 JAW11453.1 GBBI01001172 JAC17540.1 GECZ01019288 JAS50481.1 APGK01031799 APGK01031800 KB740824 ENN78837.1 GFDL01014915 JAV20130.1 DS231958 EDS29048.1 CH477487 EAT40124.1 KB632314 ERL92238.1 JXUM01028000 KQ560807 KXJ80807.1 PYGN01000174 PSN52092.1 KQ971312 EEZ98075.1 GFDF01008452 JAV05632.1 AAZX01000987 UFQT01000022 SSX18016.1 GEDC01021988 JAS15310.1 GEZM01041321 GEZM01041320 JAV80515.1 GBYB01006119 JAG75886.1 CP012526 ALC46729.1 KQ459582 KPI98744.1 KK107078 EZA60491.1 CH902617 EDV43470.1 CH940650 EDW67011.1 CH964272 EDW85086.1 CH954182 EDV53568.1 AE014297 AY119248 AAF56565.1 AAM51108.1 ALI30641.1 CH916369 EDV93698.1 CH933806 EDW15215.1 CM000364 EDX14527.1 KU659911 AOG17710.1 KQ414620 KOC67652.1 CH480828 EDW45089.1 CM000160 EDW98651.1 KQ982409 KYQ56818.1 GL440570 EFN65783.1 CM000070 EAL28307.1 CH479182 EDW33987.1 CVRI01000004 CRK87527.1 JXJN01013032 KQ981693 KYN37871.1 AAAB01008978 EAA13617.4 KQ434787 KZC05345.1 ADTU01018960 KQ980886 KYN11837.1 GGFL01004536 MBW68714.1 KQ977649 KYN00739.1 JR045824 AEY60064.1 NNAY01001960 OXU22446.1 KQ760551 OAD59820.1 GBHO01018153 GDHC01016527 GDHC01013376 JAG25451.1 JAQ02102.1 JAQ05253.1 GAMD01003219 JAA98371.1 GL888613 EGI58990.1 OUUW01000023 SPP89565.1 KQ435811 KOX72896.1 KU659988 AOG17786.1 GDIP01235789 JAI87612.1 GGMS01001633 MBY70836.1 GDIP01127358 JAL76356.1 AAZO01007496 DS235885 EEB20359.1 QOIP01000011 RLU17220.1 GDIQ01056923 JAN37814.1 CCAG010012732 KZ288285 PBC29475.1 GGMR01009519 MBY22138.1 GFXV01001046 MBW12851.1 GDIQ01086408 JAN08329.1 AXCN02000286 GL453506 EFN75929.1 KQ976439 KYM86709.1 GL732571 EFX75994.1 GDRN01029702 GDRN01029701 JAI67589.1 NEVH01011202 PNF31726.1 AJVK01019160 GDIP01115761 JAL87953.1 GDIP01188298 JAJ35104.1 ACPB03009370 KK852771 KDR16889.1 GEFM01005188 JAP70608.1 JH431874
Proteomes
UP000007151
UP000218220
UP000005204
UP000283053
UP000037069
UP000095300
+ More
UP000095301 UP000019118 UP000002320 UP000008820 UP000030742 UP000069940 UP000249989 UP000245037 UP000007266 UP000002358 UP000091820 UP000092553 UP000053268 UP000053097 UP000192221 UP000007801 UP000008792 UP000007798 UP000008711 UP000000803 UP000001070 UP000009192 UP000000304 UP000053825 UP000001292 UP000002282 UP000075809 UP000076408 UP000000311 UP000001819 UP000008744 UP000183832 UP000092443 UP000092460 UP000078541 UP000007062 UP000078200 UP000076502 UP000005205 UP000078492 UP000078542 UP000075880 UP000215335 UP000007755 UP000005203 UP000268350 UP000053105 UP000009046 UP000279307 UP000092444 UP000242457 UP000075886 UP000008237 UP000078540 UP000000305 UP000235965 UP000092462 UP000085678 UP000015103 UP000027135
UP000095301 UP000019118 UP000002320 UP000008820 UP000030742 UP000069940 UP000249989 UP000245037 UP000007266 UP000002358 UP000091820 UP000092553 UP000053268 UP000053097 UP000192221 UP000007801 UP000008792 UP000007798 UP000008711 UP000000803 UP000001070 UP000009192 UP000000304 UP000053825 UP000001292 UP000002282 UP000075809 UP000076408 UP000000311 UP000001819 UP000008744 UP000183832 UP000092443 UP000092460 UP000078541 UP000007062 UP000078200 UP000076502 UP000005205 UP000078492 UP000078542 UP000075880 UP000215335 UP000007755 UP000005203 UP000268350 UP000053105 UP000009046 UP000279307 UP000092444 UP000242457 UP000075886 UP000008237 UP000078540 UP000000305 UP000235965 UP000092462 UP000085678 UP000015103 UP000027135
Interpro
Gene 3D
ProteinModelPortal
A0A212ES41
A0A2A4JXF9
A0A2W1BZ81
H9JL06
A0A3S2M7R5
A0A2H1WYW7
+ More
A0A0L0C3C8 W8BQM5 A0A0K8VKE8 R4FND9 A0A0P4VVB4 A0A224XTM2 A0A023F8C2 A0A1I8PCD6 A0A1I8MBH5 A0A1B6FJX9 N6UJJ4 A0A1Q3EXW5 B0WJB3 Q16ZN4 U4UIK8 A0A182H750 A0A2P8Z6H3 D6WA21 A0A1L8DGP9 K7J3Z8 A0A336LJD7 A0A1B6CPE4 A0A1Y1M862 A0A0C9QQW2 A0A1A9WF52 A0A0M3QXZ4 A0A194Q178 A0A026WXM3 A0A1W4V7K6 B3M2U0 B4LYT4 B4NK44 B3P698 Q9VBH7 B4JGK1 B4K6E2 B4QW28 A0A1B3PDB5 A0A0L7R9V6 B4ICJ3 B4PSW4 A0A151X951 A0A182XXM9 E2ALA4 Q296Z9 B4GEF2 A0A1J1HHM2 A0A1A9XEX3 A0A1B0BEV8 A0A195FBW0 Q7Q294 A0A1A9VDZ9 A0A154P0A2 A0A158NKQ3 A0A195DG69 A0A2M4CTN8 A0A195CKY4 A0A182JCP4 V9IIE3 A0A232EVS8 A0A310SNV1 A0A0A9Y1D5 T1DPM6 F4X3G2 A0A088AD89 A0A3B0K611 A0A0M8ZYV7 A0A1B3PDJ1 A0A0P4Y650 A0A2S2PZ83 A0A0P5TII0 E0W403 A0A3L8D9N4 A0A0P6EWY4 A0A1B0GF17 A0A2A3ECL0 A0A2S2NYU5 A0A2H8TFL0 A0A0P6D1T9 A0A182QCC9 E2C811 A0A195BLV4 E9GX16 A0A0P4WSQ9 A0A2J7QT15 A0A1B0DQK6 A0A0P5UAJ4 A0A1S3H9R5 A0A0P5BIN1 T1HIF1 A0A067RDJ5 A0A131XWK8 T1J699 A0A1S3H7W8
A0A0L0C3C8 W8BQM5 A0A0K8VKE8 R4FND9 A0A0P4VVB4 A0A224XTM2 A0A023F8C2 A0A1I8PCD6 A0A1I8MBH5 A0A1B6FJX9 N6UJJ4 A0A1Q3EXW5 B0WJB3 Q16ZN4 U4UIK8 A0A182H750 A0A2P8Z6H3 D6WA21 A0A1L8DGP9 K7J3Z8 A0A336LJD7 A0A1B6CPE4 A0A1Y1M862 A0A0C9QQW2 A0A1A9WF52 A0A0M3QXZ4 A0A194Q178 A0A026WXM3 A0A1W4V7K6 B3M2U0 B4LYT4 B4NK44 B3P698 Q9VBH7 B4JGK1 B4K6E2 B4QW28 A0A1B3PDB5 A0A0L7R9V6 B4ICJ3 B4PSW4 A0A151X951 A0A182XXM9 E2ALA4 Q296Z9 B4GEF2 A0A1J1HHM2 A0A1A9XEX3 A0A1B0BEV8 A0A195FBW0 Q7Q294 A0A1A9VDZ9 A0A154P0A2 A0A158NKQ3 A0A195DG69 A0A2M4CTN8 A0A195CKY4 A0A182JCP4 V9IIE3 A0A232EVS8 A0A310SNV1 A0A0A9Y1D5 T1DPM6 F4X3G2 A0A088AD89 A0A3B0K611 A0A0M8ZYV7 A0A1B3PDJ1 A0A0P4Y650 A0A2S2PZ83 A0A0P5TII0 E0W403 A0A3L8D9N4 A0A0P6EWY4 A0A1B0GF17 A0A2A3ECL0 A0A2S2NYU5 A0A2H8TFL0 A0A0P6D1T9 A0A182QCC9 E2C811 A0A195BLV4 E9GX16 A0A0P4WSQ9 A0A2J7QT15 A0A1B0DQK6 A0A0P5UAJ4 A0A1S3H9R5 A0A0P5BIN1 T1HIF1 A0A067RDJ5 A0A131XWK8 T1J699 A0A1S3H7W8
PDB
5CD1
E-value=2.13422e-65,
Score=630
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
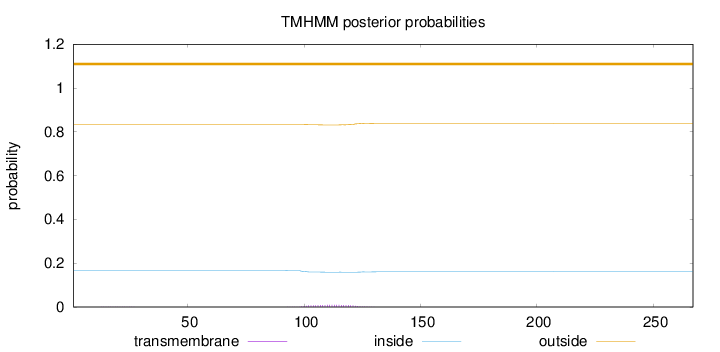
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.21331
Exp number, first 60 AAs:
0.00429
Total prob of N-in:
0.16649
outside
1 - 267
Population Genetic Test Statistics
Pi
26.004821
Theta
193.416738
Tajima's D
0.842629
CLR
0.159218
CSRT
0.615369231538423
Interpretation
Uncertain
