Pre Gene Modal
BGIBMGA010205
Annotation
PREDICTED:_vesicle-associated_membrane_protein_2-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.807 Nuclear Reliability : 1.171
Sequence
CDS
ATGAAGAGTCAGTATTCGTGTGAGACAAGGAAAATGGCAGCAGAAGGTGGCAATCCAGGTGGTCTGGCACCAGCAGAGCCCCAGACAGGTCCTGATGGAGAGATTATTGGAGGACCTCGTACACCTCAGCAGATTGCTGCTCAGAGGAGGCTTCAGCAGACACAAGCACAAGTTGATGAGGTGGTGGATATTATGAAGTCGAATGTAGATAAAGTTTTGGACAGGGACGCGAAGTTATCAGAATTGGACTACAGGGCAGATGCTCTTCAGGAAGGGGCATCGCAGTTTGAGAGACAAGCTAAGAGTCTGAAAAACAAATTTTGGTTACAGAATCTAAAGATGATAATAATAATGGGTGTGATTGGTGTTATTATCATAGGGCTGCTATTCGGAAAGTACATGTAA
Protein
MKSQYSCETRKMAAEGGNPGGLAPAEPQTGPDGEIIGGPRTPQQIAAQRRLQQTQAQVDEVVDIMKSNVDKVLDRDAKLSELDYRADALQEGASQFERQAKSLKNKFWLQNLKMIIIMGVIGVIIIGLLFGKYM
Summary
Uniprot
H9JL04
A0A212EZG6
A0A194RPH8
A0A194Q5S3
A0A0L7L7X4
A0A2W1BUG9
+ More
A0A2A4J8J8 A0A2H1X1V3 A0A3S2LTB3 A0A1J1IRW0 A0A0A9WWU5 A0A0A9WPG0 E0W1F5 A0A1L8DVG7 T1E1U4 A0A1B0DBZ6 A0A336JXK9 A0A1Q3G578 U5EXV1 A0A1B0CIW1 A0A2P8YPR5 A0A1B6DLT3 A0A182SJS7 A0A1Q3FVI5 A0A067RVI7 A0A1B6E907 A0A182GF13 A0A182Y0X9 A0A0P6IUI5 B0WRV6 A0A1Q3FRM4 A0A0C9S1F2 A0A182J4H7 A0A0C9RMG5 A0A2H1VK35 B0XAS2 A0A1Q3FVI9 A0A2A4K3R5 A0A2A4K214 A0A2J7RF77 A0A2S2P093 A0A3Q0IXP7 A0A194PZJ9 A0A194RK44 F5HSC4 W5JQA4 A0A1B6M4V0 A0A1B6F509 A0A1B6HS35 B4N705 A0A0Q9X2X7
A0A2A4J8J8 A0A2H1X1V3 A0A3S2LTB3 A0A1J1IRW0 A0A0A9WWU5 A0A0A9WPG0 E0W1F5 A0A1L8DVG7 T1E1U4 A0A1B0DBZ6 A0A336JXK9 A0A1Q3G578 U5EXV1 A0A1B0CIW1 A0A2P8YPR5 A0A1B6DLT3 A0A182SJS7 A0A1Q3FVI5 A0A067RVI7 A0A1B6E907 A0A182GF13 A0A182Y0X9 A0A0P6IUI5 B0WRV6 A0A1Q3FRM4 A0A0C9S1F2 A0A182J4H7 A0A0C9RMG5 A0A2H1VK35 B0XAS2 A0A1Q3FVI9 A0A2A4K3R5 A0A2A4K214 A0A2J7RF77 A0A2S2P093 A0A3Q0IXP7 A0A194PZJ9 A0A194RK44 F5HSC4 W5JQA4 A0A1B6M4V0 A0A1B6F509 A0A1B6HS35 B4N705 A0A0Q9X2X7
Pubmed
EMBL
BABH01011423
AGBW02011314
OWR46834.1
KQ460075
KPJ17926.1
KQ459582
+ More
KPI98750.1 JTDY01002426 KOB71439.1 KZ149914 PZC78001.1 NWSH01002524 PCG68086.1 ODYU01012811 SOQ59303.1 RSAL01000013 RVE53389.1 CVRI01000057 CRL02292.1 GBHO01034274 GBHO01034272 JAG09330.1 JAG09332.1 GBHO01034273 GBHO01034271 GBRD01012854 GDHC01017411 GDHC01009745 JAG09331.1 JAG09333.1 JAG52972.1 JAQ01218.1 JAQ08884.1 DS235870 EEB19461.1 GFDF01003664 JAV10420.1 GALA01001359 JAA93493.1 AJVK01013865 UFQS01000016 UFQT01000016 SSW97404.1 SSX17790.1 GFDL01000084 JAV34961.1 GANO01000737 JAB59134.1 AJWK01013814 AJWK01013815 AJWK01013816 AJWK01013817 PYGN01000445 PSN46241.1 GEDC01010690 JAS26608.1 GFDL01003448 JAV31597.1 KK852439 KDR23889.1 GEDC01002923 JAS34375.1 JXUM01059063 KQ562033 KXJ76854.1 GDUN01001110 JAN94809.1 DS232061 EDS33521.1 GFDL01004828 JAV30217.1 GBYB01014601 JAG84368.1 GBYB01014602 JAG84369.1 ODYU01002831 SOQ40802.1 DS232594 EDS43861.1 GFDL01003583 JAV31462.1 NWSH01000216 PCG78310.1 PCG78311.1 NEVH01004413 PNF39470.1 GGMR01010274 MBY22893.1 KPI98757.1 KPJ17917.1 AB630175 BAK23255.1 ADMH02000483 ETN66311.1 GEBQ01009028 JAT30949.1 GECZ01024806 JAS44963.1 GECU01030212 JAS77494.1 CH964168 EDW80144.1 KRF99245.1
KPI98750.1 JTDY01002426 KOB71439.1 KZ149914 PZC78001.1 NWSH01002524 PCG68086.1 ODYU01012811 SOQ59303.1 RSAL01000013 RVE53389.1 CVRI01000057 CRL02292.1 GBHO01034274 GBHO01034272 JAG09330.1 JAG09332.1 GBHO01034273 GBHO01034271 GBRD01012854 GDHC01017411 GDHC01009745 JAG09331.1 JAG09333.1 JAG52972.1 JAQ01218.1 JAQ08884.1 DS235870 EEB19461.1 GFDF01003664 JAV10420.1 GALA01001359 JAA93493.1 AJVK01013865 UFQS01000016 UFQT01000016 SSW97404.1 SSX17790.1 GFDL01000084 JAV34961.1 GANO01000737 JAB59134.1 AJWK01013814 AJWK01013815 AJWK01013816 AJWK01013817 PYGN01000445 PSN46241.1 GEDC01010690 JAS26608.1 GFDL01003448 JAV31597.1 KK852439 KDR23889.1 GEDC01002923 JAS34375.1 JXUM01059063 KQ562033 KXJ76854.1 GDUN01001110 JAN94809.1 DS232061 EDS33521.1 GFDL01004828 JAV30217.1 GBYB01014601 JAG84368.1 GBYB01014602 JAG84369.1 ODYU01002831 SOQ40802.1 DS232594 EDS43861.1 GFDL01003583 JAV31462.1 NWSH01000216 PCG78310.1 PCG78311.1 NEVH01004413 PNF39470.1 GGMR01010274 MBY22893.1 KPI98757.1 KPJ17917.1 AB630175 BAK23255.1 ADMH02000483 ETN66311.1 GEBQ01009028 JAT30949.1 GECZ01024806 JAS44963.1 GECU01030212 JAS77494.1 CH964168 EDW80144.1 KRF99245.1
Proteomes
PRIDE
Pfam
PF00957 Synaptobrevin
ProteinModelPortal
H9JL04
A0A212EZG6
A0A194RPH8
A0A194Q5S3
A0A0L7L7X4
A0A2W1BUG9
+ More
A0A2A4J8J8 A0A2H1X1V3 A0A3S2LTB3 A0A1J1IRW0 A0A0A9WWU5 A0A0A9WPG0 E0W1F5 A0A1L8DVG7 T1E1U4 A0A1B0DBZ6 A0A336JXK9 A0A1Q3G578 U5EXV1 A0A1B0CIW1 A0A2P8YPR5 A0A1B6DLT3 A0A182SJS7 A0A1Q3FVI5 A0A067RVI7 A0A1B6E907 A0A182GF13 A0A182Y0X9 A0A0P6IUI5 B0WRV6 A0A1Q3FRM4 A0A0C9S1F2 A0A182J4H7 A0A0C9RMG5 A0A2H1VK35 B0XAS2 A0A1Q3FVI9 A0A2A4K3R5 A0A2A4K214 A0A2J7RF77 A0A2S2P093 A0A3Q0IXP7 A0A194PZJ9 A0A194RK44 F5HSC4 W5JQA4 A0A1B6M4V0 A0A1B6F509 A0A1B6HS35 B4N705 A0A0Q9X2X7
A0A2A4J8J8 A0A2H1X1V3 A0A3S2LTB3 A0A1J1IRW0 A0A0A9WWU5 A0A0A9WPG0 E0W1F5 A0A1L8DVG7 T1E1U4 A0A1B0DBZ6 A0A336JXK9 A0A1Q3G578 U5EXV1 A0A1B0CIW1 A0A2P8YPR5 A0A1B6DLT3 A0A182SJS7 A0A1Q3FVI5 A0A067RVI7 A0A1B6E907 A0A182GF13 A0A182Y0X9 A0A0P6IUI5 B0WRV6 A0A1Q3FRM4 A0A0C9S1F2 A0A182J4H7 A0A0C9RMG5 A0A2H1VK35 B0XAS2 A0A1Q3FVI9 A0A2A4K3R5 A0A2A4K214 A0A2J7RF77 A0A2S2P093 A0A3Q0IXP7 A0A194PZJ9 A0A194RK44 F5HSC4 W5JQA4 A0A1B6M4V0 A0A1B6F509 A0A1B6HS35 B4N705 A0A0Q9X2X7
PDB
2KOG
E-value=1.19333e-16,
Score=205
Ontologies
GO
Topology
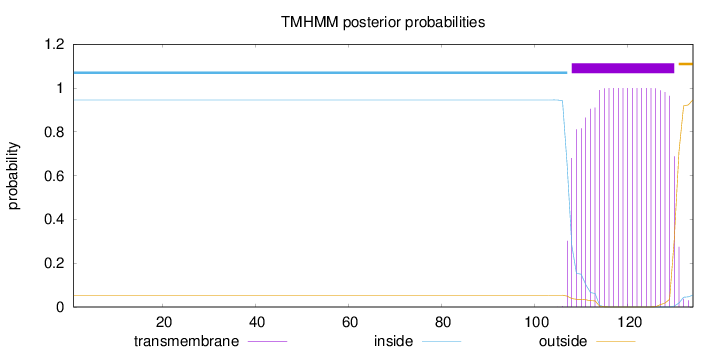
Length:
134
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.24734
Exp number, first 60 AAs:
0
Total prob of N-in:
0.94591
inside
1 - 107
TMhelix
108 - 130
outside
131 - 134
Population Genetic Test Statistics
Pi
326.709405
Theta
185.528639
Tajima's D
2.276271
CLR
0.194729
CSRT
0.919154042297885
Interpretation
Uncertain
