Gene
KWMTBOMO03873 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010199
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_FKBP2_[Amyelois_transitella]
Full name
Peptidylprolyl isomerase
Location in the cell
Cytoplasmic Reliability : 1.419 Mitochondrial Reliability : 1.465 Nuclear Reliability : 1.128
Sequence
CDS
ATGTTTAAGTTAGTGGTAATAGAAAGTACAGCTTCAAAGAAACTACAAATTGGCATTAAGAAAAGACCTGAGGACTGTTCCATCAAGAGCCGCAAAGGAGATCTTTTGCATATGCACTACACTGGCACTTTAGATGACGGTACAGAATTTGATAGTTCAATTCCTCGAGGTAACCCCTTAACCTTCAAACTTGGTTCTGGTCAAGTTATTAAGGGTTGGGATCAAGGGCTACTTGGAATGTGCGAAGGAGAACAACGTAAACTTGTCATTCCACCGGAATTGGCTTATGGTTCAGCTGGAGCACCACCAAAGATACCAAAGTCTGCTACTCTTACTTTTCACGTAGAATTGGTGAAAATTGAAAGAAAAGATGAATTATAA
Protein
MFKLVVIESTASKKLQIGIKKRPEDCSIKSRKGDLLHMHYTGTLDDGTEFDSSIPRGNPLTFKLGSGQVIKGWDQGLLGMCEGEQRKLVIPPELAYGSAGAPPKIPKSATLTFHVELVKIERKDEL
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Feature
chain Peptidylprolyl isomerase
Uniprot
H9JKZ8
A0A2W1BSH3
A0A2H1X2I0
A0A2A4K2L9
I4DKC7
A0A194PZL2
+ More
A0A212FFJ5 A0A194RJA8 S4PDJ4 A0A0N8E7X3 A0A0P6AE28 E9FTU8 A0A164M3C1 A0A0P5FS36 A0A0P5WQ80 E0VWX3 A0A2R5LJ11 A0A1Z5KWH5 A0A147B9V8 Q7ZYC8 A0A182QPG0 A0A023FWG3 T1FAA8 A0A0L7L7K5 A0A023FNR3 A0A182XE97 A0A182V638 A0A182LC75 A0A182U6X3 A0A182HJ24 A0A224XZI3 T1HIE0 A0A0S7JXK2 Q7QBK5 A0A293N324 B7P5F5 E3TDD6 A0A1B6C5K0 S4RI03 R7UCZ7 E3TDS0 A0A131Z1L4 A0A182PSX0 L7M243 A0A182RYC9 A0A023GMI2 A0A182JS62 A0A182NSD3 A0A1A8S8U8 A0A1A8KJI9 A0A1A8UFX4 A0A161MBF5 A0A1A8RP79 G3NSC6 A0A1S3IHV9 A0A1A8CDQ5 G3NSC0 A0A0C9SDN2 A0A1E1X1J3 A0A0E9XQJ5 A0A1A8FV51 A9JSW0 Q6GQ21 A0A067RAY6 A9ULB7 A0A182W2T9 A0A3Q3VUV7 A0A182XVK4 A0A1A8QTC5 A0A084WSZ1 A0A1A8DNL7 A0A182T5V2 A0A3N0YNB3 A0A3B4C5W3 A0A087YRT9 A0A3B3UZ49 A0A1B6GA79 A0A224YYW8 A0A1B6JDN5 H3C2C4 A0A0K8R9J8 A0A3Q1B3H6 A0A3P8T2Z0 U3E6G2 W5KIW4 A0A3Q2V5X6 A0A3P9B8C0 Q16Z09 C1BM14 U3CBQ7 I3JQV6 A0A0P7VS48 A0A087TFA4 A0A2I4BZT0 A0A182J4A2 A0A2M3ZAT9 T1D9Q6 V9LIL1 V9LIM0 A0A1A7Z2Z8
A0A212FFJ5 A0A194RJA8 S4PDJ4 A0A0N8E7X3 A0A0P6AE28 E9FTU8 A0A164M3C1 A0A0P5FS36 A0A0P5WQ80 E0VWX3 A0A2R5LJ11 A0A1Z5KWH5 A0A147B9V8 Q7ZYC8 A0A182QPG0 A0A023FWG3 T1FAA8 A0A0L7L7K5 A0A023FNR3 A0A182XE97 A0A182V638 A0A182LC75 A0A182U6X3 A0A182HJ24 A0A224XZI3 T1HIE0 A0A0S7JXK2 Q7QBK5 A0A293N324 B7P5F5 E3TDD6 A0A1B6C5K0 S4RI03 R7UCZ7 E3TDS0 A0A131Z1L4 A0A182PSX0 L7M243 A0A182RYC9 A0A023GMI2 A0A182JS62 A0A182NSD3 A0A1A8S8U8 A0A1A8KJI9 A0A1A8UFX4 A0A161MBF5 A0A1A8RP79 G3NSC6 A0A1S3IHV9 A0A1A8CDQ5 G3NSC0 A0A0C9SDN2 A0A1E1X1J3 A0A0E9XQJ5 A0A1A8FV51 A9JSW0 Q6GQ21 A0A067RAY6 A9ULB7 A0A182W2T9 A0A3Q3VUV7 A0A182XVK4 A0A1A8QTC5 A0A084WSZ1 A0A1A8DNL7 A0A182T5V2 A0A3N0YNB3 A0A3B4C5W3 A0A087YRT9 A0A3B3UZ49 A0A1B6GA79 A0A224YYW8 A0A1B6JDN5 H3C2C4 A0A0K8R9J8 A0A3Q1B3H6 A0A3P8T2Z0 U3E6G2 W5KIW4 A0A3Q2V5X6 A0A3P9B8C0 Q16Z09 C1BM14 U3CBQ7 I3JQV6 A0A0P7VS48 A0A087TFA4 A0A2I4BZT0 A0A182J4A2 A0A2M3ZAT9 T1D9Q6 V9LIL1 V9LIM0 A0A1A7Z2Z8
EC Number
5.2.1.8
Pubmed
19121390
28756777
22651552
26354079
22118469
23622113
+ More
21292972 20566863 28528879 27762356 23254933 26227816 20966253 12364791 14747013 17210077 20634964 26830274 25576852 26131772 28503490 25613341 23594743 24845553 20431018 25244985 24438588 28797301 15496914 25243066 25329095 25186727 17510324 23758969 24402279
21292972 20566863 28528879 27762356 23254933 26227816 20966253 12364791 14747013 17210077 20634964 26830274 25576852 26131772 28503490 25613341 23594743 24845553 20431018 25244985 24438588 28797301 15496914 25243066 25329095 25186727 17510324 23758969 24402279
EMBL
BABH01011400
KZ149914
PZC78018.1
ODYU01012499
SOQ58844.1
NWSH01000216
+ More
PCG78299.1 AK401745 BAM18367.1 KQ459582 KPI98761.1 AGBW02008814 OWR52502.1 KQ460075 KPJ17913.1 GAIX01003651 JAA88909.1 GDIQ01053084 JAN41653.1 GDIP01031032 JAM72683.1 GL732524 EFX89578.1 LRGB01003101 KZS04687.1 GDIP01144462 JAJ78940.1 GDIP01083869 JAM19846.1 DS235824 EEB17879.1 GGLE01005406 MBY09532.1 GFJQ02007504 JAV99465.1 GEIB01000778 JAR87222.1 BC043844 BC108494 CM004472 AAH43844.1 AAI08495.1 OCT84072.1 AXCN02000238 GBBL01002247 JAC25073.1 AMQM01005587 KB097026 ESN99916.1 JTDY01002505 KOB71271.1 GBBK01001997 JAC22485.1 APCN01002118 GFTR01002404 JAW14022.1 ACPB03009369 GBYX01226753 GBYX01226752 JAO69916.1 AAAB01008879 EAA08436.4 GFWV01022735 MAA47462.1 ABJB010135641 ABJB010331434 ABJB010475883 ABJB010831436 ABJB010896591 DS640514 EEC01827.1 GU588366 ADO28322.1 GEDC01028614 JAS08684.1 AMQN01001773 KB305378 ELU01137.1 GU588500 ADO28456.1 GEDV01004526 JAP84031.1 GACK01007836 JAA57198.1 GBBM01000373 GBBM01000372 JAC35046.1 HAEI01012218 SBS14687.1 HAED01003501 HAEE01012393 SBR32443.1 HADY01018677 HAEJ01006064 SBS46521.1 GEMB01006383 JAR96955.1 HAEH01018586 SBS07915.1 HADZ01012970 SBP76911.1 GBZX01000578 JAG92162.1 GFAC01006045 JAT93143.1 GBXM01003866 JAI04712.1 HAEB01016753 HAEC01007129 SBQ63280.1 BX465183 BC155099 AAI55100.1 BC072927 CM004473 AAH72927.1 OCT81721.1 KK852771 KDR16890.1 AAMC01101476 BC157195 BC171197 BC171199 AAI57196.1 AAI71197.1 HAEF01021175 HAEG01014026 SBR96508.1 ATLV01026773 KE525419 KFB53335.1 HAEA01007127 SBQ35607.1 RJVU01035392 ROL47689.1 AYCK01020911 GECZ01010437 JAS59332.1 GFPF01007814 MAA18960.1 GECU01010643 JAS97063.1 GADI01005988 JAA67820.1 GAMT01007049 GAMT01007048 GAMQ01002057 GAMQ01002056 GAMP01006262 GAMP01006261 JAB04812.1 JAB39794.1 JAB46493.1 CH477505 EAT39857.1 BT075643 ACO10067.1 GAMS01007383 GAMS01007382 GAMS01007381 JAB15754.1 AERX01028214 AERX01028215 JARO02000244 KPP79274.1 KK114963 KFM63793.1 AXCP01008524 GGFM01004905 MBW25656.1 GAAZ01001854 JAA96089.1 JW880688 AFP13205.1 JW880938 AFP13455.1 HADW01004265 HADX01014985 SBP37217.1
PCG78299.1 AK401745 BAM18367.1 KQ459582 KPI98761.1 AGBW02008814 OWR52502.1 KQ460075 KPJ17913.1 GAIX01003651 JAA88909.1 GDIQ01053084 JAN41653.1 GDIP01031032 JAM72683.1 GL732524 EFX89578.1 LRGB01003101 KZS04687.1 GDIP01144462 JAJ78940.1 GDIP01083869 JAM19846.1 DS235824 EEB17879.1 GGLE01005406 MBY09532.1 GFJQ02007504 JAV99465.1 GEIB01000778 JAR87222.1 BC043844 BC108494 CM004472 AAH43844.1 AAI08495.1 OCT84072.1 AXCN02000238 GBBL01002247 JAC25073.1 AMQM01005587 KB097026 ESN99916.1 JTDY01002505 KOB71271.1 GBBK01001997 JAC22485.1 APCN01002118 GFTR01002404 JAW14022.1 ACPB03009369 GBYX01226753 GBYX01226752 JAO69916.1 AAAB01008879 EAA08436.4 GFWV01022735 MAA47462.1 ABJB010135641 ABJB010331434 ABJB010475883 ABJB010831436 ABJB010896591 DS640514 EEC01827.1 GU588366 ADO28322.1 GEDC01028614 JAS08684.1 AMQN01001773 KB305378 ELU01137.1 GU588500 ADO28456.1 GEDV01004526 JAP84031.1 GACK01007836 JAA57198.1 GBBM01000373 GBBM01000372 JAC35046.1 HAEI01012218 SBS14687.1 HAED01003501 HAEE01012393 SBR32443.1 HADY01018677 HAEJ01006064 SBS46521.1 GEMB01006383 JAR96955.1 HAEH01018586 SBS07915.1 HADZ01012970 SBP76911.1 GBZX01000578 JAG92162.1 GFAC01006045 JAT93143.1 GBXM01003866 JAI04712.1 HAEB01016753 HAEC01007129 SBQ63280.1 BX465183 BC155099 AAI55100.1 BC072927 CM004473 AAH72927.1 OCT81721.1 KK852771 KDR16890.1 AAMC01101476 BC157195 BC171197 BC171199 AAI57196.1 AAI71197.1 HAEF01021175 HAEG01014026 SBR96508.1 ATLV01026773 KE525419 KFB53335.1 HAEA01007127 SBQ35607.1 RJVU01035392 ROL47689.1 AYCK01020911 GECZ01010437 JAS59332.1 GFPF01007814 MAA18960.1 GECU01010643 JAS97063.1 GADI01005988 JAA67820.1 GAMT01007049 GAMT01007048 GAMQ01002057 GAMQ01002056 GAMP01006262 GAMP01006261 JAB04812.1 JAB39794.1 JAB46493.1 CH477505 EAT39857.1 BT075643 ACO10067.1 GAMS01007383 GAMS01007382 GAMS01007381 JAB15754.1 AERX01028214 AERX01028215 JARO02000244 KPP79274.1 KK114963 KFM63793.1 AXCP01008524 GGFM01004905 MBW25656.1 GAAZ01001854 JAA96089.1 JW880688 AFP13205.1 JW880938 AFP13455.1 HADW01004265 HADX01014985 SBP37217.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000000305
+ More
UP000076858 UP000009046 UP000186698 UP000075886 UP000015101 UP000037510 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000015103 UP000007062 UP000001555 UP000245300 UP000014760 UP000221080 UP000075885 UP000075900 UP000075881 UP000075884 UP000007635 UP000085678 UP000000437 UP000027135 UP000008143 UP000075920 UP000261620 UP000076408 UP000030765 UP000075901 UP000261440 UP000028760 UP000261500 UP000007303 UP000257160 UP000265080 UP000008225 UP000018467 UP000264840 UP000265160 UP000008820 UP000005207 UP000034805 UP000054359 UP000192220 UP000075880
UP000076858 UP000009046 UP000186698 UP000075886 UP000015101 UP000037510 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000015103 UP000007062 UP000001555 UP000245300 UP000014760 UP000221080 UP000075885 UP000075900 UP000075881 UP000075884 UP000007635 UP000085678 UP000000437 UP000027135 UP000008143 UP000075920 UP000261620 UP000076408 UP000030765 UP000075901 UP000261440 UP000028760 UP000261500 UP000007303 UP000257160 UP000265080 UP000008225 UP000018467 UP000264840 UP000265160 UP000008820 UP000005207 UP000034805 UP000054359 UP000192220 UP000075880
Pfam
PF00254 FKBP_C
Interpro
IPR001179
PPIase_FKBP_dom
ProteinModelPortal
H9JKZ8
A0A2W1BSH3
A0A2H1X2I0
A0A2A4K2L9
I4DKC7
A0A194PZL2
+ More
A0A212FFJ5 A0A194RJA8 S4PDJ4 A0A0N8E7X3 A0A0P6AE28 E9FTU8 A0A164M3C1 A0A0P5FS36 A0A0P5WQ80 E0VWX3 A0A2R5LJ11 A0A1Z5KWH5 A0A147B9V8 Q7ZYC8 A0A182QPG0 A0A023FWG3 T1FAA8 A0A0L7L7K5 A0A023FNR3 A0A182XE97 A0A182V638 A0A182LC75 A0A182U6X3 A0A182HJ24 A0A224XZI3 T1HIE0 A0A0S7JXK2 Q7QBK5 A0A293N324 B7P5F5 E3TDD6 A0A1B6C5K0 S4RI03 R7UCZ7 E3TDS0 A0A131Z1L4 A0A182PSX0 L7M243 A0A182RYC9 A0A023GMI2 A0A182JS62 A0A182NSD3 A0A1A8S8U8 A0A1A8KJI9 A0A1A8UFX4 A0A161MBF5 A0A1A8RP79 G3NSC6 A0A1S3IHV9 A0A1A8CDQ5 G3NSC0 A0A0C9SDN2 A0A1E1X1J3 A0A0E9XQJ5 A0A1A8FV51 A9JSW0 Q6GQ21 A0A067RAY6 A9ULB7 A0A182W2T9 A0A3Q3VUV7 A0A182XVK4 A0A1A8QTC5 A0A084WSZ1 A0A1A8DNL7 A0A182T5V2 A0A3N0YNB3 A0A3B4C5W3 A0A087YRT9 A0A3B3UZ49 A0A1B6GA79 A0A224YYW8 A0A1B6JDN5 H3C2C4 A0A0K8R9J8 A0A3Q1B3H6 A0A3P8T2Z0 U3E6G2 W5KIW4 A0A3Q2V5X6 A0A3P9B8C0 Q16Z09 C1BM14 U3CBQ7 I3JQV6 A0A0P7VS48 A0A087TFA4 A0A2I4BZT0 A0A182J4A2 A0A2M3ZAT9 T1D9Q6 V9LIL1 V9LIM0 A0A1A7Z2Z8
A0A212FFJ5 A0A194RJA8 S4PDJ4 A0A0N8E7X3 A0A0P6AE28 E9FTU8 A0A164M3C1 A0A0P5FS36 A0A0P5WQ80 E0VWX3 A0A2R5LJ11 A0A1Z5KWH5 A0A147B9V8 Q7ZYC8 A0A182QPG0 A0A023FWG3 T1FAA8 A0A0L7L7K5 A0A023FNR3 A0A182XE97 A0A182V638 A0A182LC75 A0A182U6X3 A0A182HJ24 A0A224XZI3 T1HIE0 A0A0S7JXK2 Q7QBK5 A0A293N324 B7P5F5 E3TDD6 A0A1B6C5K0 S4RI03 R7UCZ7 E3TDS0 A0A131Z1L4 A0A182PSX0 L7M243 A0A182RYC9 A0A023GMI2 A0A182JS62 A0A182NSD3 A0A1A8S8U8 A0A1A8KJI9 A0A1A8UFX4 A0A161MBF5 A0A1A8RP79 G3NSC6 A0A1S3IHV9 A0A1A8CDQ5 G3NSC0 A0A0C9SDN2 A0A1E1X1J3 A0A0E9XQJ5 A0A1A8FV51 A9JSW0 Q6GQ21 A0A067RAY6 A9ULB7 A0A182W2T9 A0A3Q3VUV7 A0A182XVK4 A0A1A8QTC5 A0A084WSZ1 A0A1A8DNL7 A0A182T5V2 A0A3N0YNB3 A0A3B4C5W3 A0A087YRT9 A0A3B3UZ49 A0A1B6GA79 A0A224YYW8 A0A1B6JDN5 H3C2C4 A0A0K8R9J8 A0A3Q1B3H6 A0A3P8T2Z0 U3E6G2 W5KIW4 A0A3Q2V5X6 A0A3P9B8C0 Q16Z09 C1BM14 U3CBQ7 I3JQV6 A0A0P7VS48 A0A087TFA4 A0A2I4BZT0 A0A182J4A2 A0A2M3ZAT9 T1D9Q6 V9LIL1 V9LIM0 A0A1A7Z2Z8
PDB
4NNR
E-value=9.55282e-46,
Score=455
Ontologies
GO
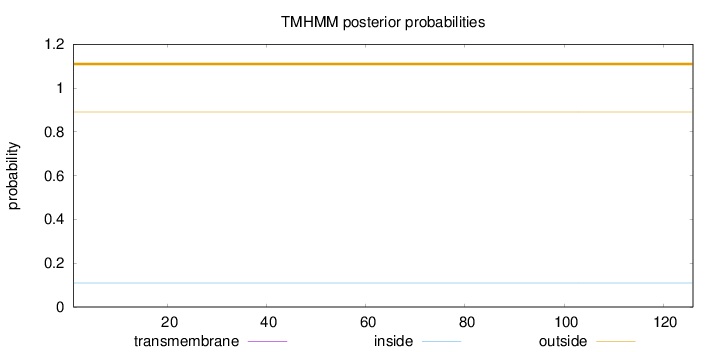
Topology
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5e-05
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10926
outside
1 - 126
Population Genetic Test Statistics
Pi
254.360535
Theta
180.2265
Tajima's D
1.290839
CLR
0
CSRT
0.734813259337033
Interpretation
Uncertain
