Pre Gene Modal
BGIBMGA010197
Annotation
PREDICTED:_short_coiled-coil_protein_homolog_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 3.175
Sequence
CDS
ATGTCCGTTATAATGCAGAAATGTGGTGATGATAATATACCTCTTGCAGATGACGACCCTCAAGTCATCATTTCCGATGACACGGATAACAACCGTTTAGATCATGGTCGTTCGATGGACTCATTACCCAGTTCTTACACTGCTGGATCTTCAAGCCCTGGCTTAAACGGACCAGCTAGTTTTGAACCTGATTCCGGCATAGATGAGCAAGAGGAAAAGGCTCGTTTGATATCACAAGTTTTGGAGCTTCAAAATACTCTGGATGACCTTTCACAAAGAGTTGACTCTGTTAAAGAAGAAAACCTAAAACTCCGCTCCGAGAATCAAGTTCTCGGTCAATATATAGAAAATCTCATGTCGGCATCATCAGTATTTCAATCCACTACTCCCAATATACAAAAGAAGTGA
Protein
MSVIMQKCGDDNIPLADDDPQVIISDDTDNNRLDHGRSMDSLPSSYTAGSSSPGLNGPASFEPDSGIDEQEEKARLISQVLELQNTLDDLSQRVDSVKEENLKLRSENQVLGQYIENLMSASSVFQSTTPNIQKK
Summary
Uniprot
H9JKZ6
S4PXK0
A0A3S2LTA3
A0A194Q5T8
A0A1E1WH64
A0A212FFJ1
+ More
A0A2H1WY60 A0A2W1BSG2 A0A2A4K4B8 A0A0L7KUN1 A0A067RCL3 D7EII5 A0A1Y1M9P2 A0A0K8TNS4 R4WRI2 A0A224XZK2 A0A0P4VS68 R4G849 J3JU93 E0W1C6 A0A1B6C8T3 A0A0A9VTK3 A0A195E5R6 F4W9Z2 A0A0A9W2F5 A0A1W4XAI2 A0A034VGK8 A0A0K8VR73 A0A1B6MRB1 A0A0A1WNH3 A0A026WRB0 A0A0L0CHZ0 A0A151HYK8 A0A158NR42 A0A182K7S3 A0A0L7QRC3 A0A151ILW5 A0A1L8EDN9 A0A154P9M7 A0A1B6HPT1 A0A1I8PFZ1 A0A1B6FGC6 W8B725 A0A2S2Q095 A0A0J7L642 A0A182ME14 A0A182PRQ8 A0A182QNT3 W5J8K2 A0A182VVW0 A0A182FRN3 A0A182RA37 U5EGH2 T1IQD3 T1PGF2 A0A2H8TN14 A0A310S530 A0A2J7R1N6 A0A151JWG4 A0A2A3ELN6 V9IJG7 A0A088AK63 A0A182TFC5 A0A182X9I2 Q7PX88 A0A182HKG5 A0A084W0M4 A0A182IK87 V5I7A6 E9IPP4 J9JLL5 A0A182URT8 A0A182L8P9 A0A1B0G6R5 A0A1A9UTP1 A0A1A9ZTB8 A0A1B0B3S5 A0A1A9YK27 Q17D62 A0A023EDY1 A0A1W4UR88 A0A0K8T6F9 T1DRA7 A0A224Y8N3 L7M3M5 A0A131YIE3 A0A131XBM5 A0A1Q3F5J6 B4NT73 B4PRS5 B4IC74 B3P629 Q9VB51 K7ISK8 A0A1E1XQ16 F0J9V3 A0A0C9QZM7 B0X1C6 B4K781
A0A2H1WY60 A0A2W1BSG2 A0A2A4K4B8 A0A0L7KUN1 A0A067RCL3 D7EII5 A0A1Y1M9P2 A0A0K8TNS4 R4WRI2 A0A224XZK2 A0A0P4VS68 R4G849 J3JU93 E0W1C6 A0A1B6C8T3 A0A0A9VTK3 A0A195E5R6 F4W9Z2 A0A0A9W2F5 A0A1W4XAI2 A0A034VGK8 A0A0K8VR73 A0A1B6MRB1 A0A0A1WNH3 A0A026WRB0 A0A0L0CHZ0 A0A151HYK8 A0A158NR42 A0A182K7S3 A0A0L7QRC3 A0A151ILW5 A0A1L8EDN9 A0A154P9M7 A0A1B6HPT1 A0A1I8PFZ1 A0A1B6FGC6 W8B725 A0A2S2Q095 A0A0J7L642 A0A182ME14 A0A182PRQ8 A0A182QNT3 W5J8K2 A0A182VVW0 A0A182FRN3 A0A182RA37 U5EGH2 T1IQD3 T1PGF2 A0A2H8TN14 A0A310S530 A0A2J7R1N6 A0A151JWG4 A0A2A3ELN6 V9IJG7 A0A088AK63 A0A182TFC5 A0A182X9I2 Q7PX88 A0A182HKG5 A0A084W0M4 A0A182IK87 V5I7A6 E9IPP4 J9JLL5 A0A182URT8 A0A182L8P9 A0A1B0G6R5 A0A1A9UTP1 A0A1A9ZTB8 A0A1B0B3S5 A0A1A9YK27 Q17D62 A0A023EDY1 A0A1W4UR88 A0A0K8T6F9 T1DRA7 A0A224Y8N3 L7M3M5 A0A131YIE3 A0A131XBM5 A0A1Q3F5J6 B4NT73 B4PRS5 B4IC74 B3P629 Q9VB51 K7ISK8 A0A1E1XQ16 F0J9V3 A0A0C9QZM7 B0X1C6 B4K781
Pubmed
19121390
23622113
26354079
22118469
28756777
26227816
+ More
24845553 18362917 19820115 28004739 26369729 23691247 27129103 22516182 23537049 20566863 25401762 26823975 21719571 25348373 25830018 24508170 30249741 26108605 21347285 24495485 20920257 23761445 25315136 12364791 14747013 17210077 24438588 21282665 20966253 17510324 24945155 26483478 28797301 25576852 26830274 28049606 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 29209593 21362191 26131772
24845553 18362917 19820115 28004739 26369729 23691247 27129103 22516182 23537049 20566863 25401762 26823975 21719571 25348373 25830018 24508170 30249741 26108605 21347285 24495485 20920257 23761445 25315136 12364791 14747013 17210077 24438588 21282665 20966253 17510324 24945155 26483478 28797301 25576852 26830274 28049606 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 29209593 21362191 26131772
EMBL
BABH01011399
GAIX01004903
JAA87657.1
RSAL01000013
RVE53370.1
KQ459582
+ More
KPI98765.1 GDQN01004808 JAT86246.1 AGBW02008814 OWR52498.1 ODYU01011965 SOQ58010.1 KZ149914 PZC78022.1 NWSH01000174 PCG78753.1 PCG78754.1 JTDY01005451 KOB66987.1 KK852790 KDR16503.1 DS497681 EFA12034.1 GEZM01039645 JAV81300.1 GDAI01001561 JAI16042.1 AK417297 BAN20512.1 GFTR01002366 JAW14060.1 GDKW01001826 JAI54769.1 ACPB03017123 GAHY01001592 JAA75918.1 APGK01047102 BT126806 KB741077 KB632384 AEE61768.1 ENN74130.1 ERL94261.1 DS235869 EEB19432.1 GEDC01027381 JAS09917.1 GBHO01044545 GDHC01021505 JAF99058.1 JAP97123.1 KQ979608 KYN20508.1 GL888035 EGI68982.1 GBHO01044544 GDHC01001300 JAF99059.1 JAQ17329.1 GAKP01017745 JAC41207.1 GDHF01010935 GDHF01007070 GDHF01002034 JAI41379.1 JAI45244.1 JAI50280.1 GEBQ01001529 JAT38448.1 GBXI01013925 JAD00367.1 KK107144 QOIP01000003 EZA57609.1 RLU24319.1 JRES01000378 KNC31827.1 KQ976717 KYM76783.1 ADTU01023801 ADTU01023802 KQ414782 KOC61185.1 KQ977085 KYN05881.1 GFDG01001968 JAV16831.1 KQ434849 KZC08527.1 GECU01031045 GECU01022510 GECU01019250 JAS76661.1 JAS85196.1 JAS88456.1 GECZ01020513 JAS49256.1 GAMC01009590 JAB96965.1 GGMS01001898 MBY71101.1 LBMM01000401 KMQ98442.1 AXCM01000480 AXCN02001512 ADMH02002066 ETN59713.1 GANO01003442 JAB56429.1 JH431303 KA647882 AFP62511.1 GFXV01002873 MBW14678.1 KQ772519 OAD52447.1 NEVH01008204 PNF34749.1 KQ981650 KYN38645.1 KZ288224 PBC31941.1 JR050206 AEY61200.1 AAAB01008987 EAA01728.5 APCN01000828 ATLV01019123 KE525262 KFB43768.1 AXCP01002654 GALX01006770 JAB61696.1 GL764558 EFZ17457.1 ABLF02033604 CCAG010013859 JXJN01008014 CH477299 EAT44308.1 JXUM01033474 GAPW01005785 KQ560989 JAC07813.1 KXJ80110.1 GBRD01005056 JAG60765.1 GAMD01000710 JAB00881.1 GFPF01000763 MAA11909.1 GACK01006414 JAA58620.1 GEDV01009528 JAP79029.1 GEFH01005076 JAP63505.1 GFDL01012213 JAV22832.1 CH982788 EDX15763.1 CM000160 EDW98518.1 CH480828 EDW45232.1 CH954182 EDV53429.1 AE014297 AY084197 KX531576 AAF56692.1 AAL89935.1 ANY27386.1 GFAA01002056 JAU01379.1 BK007654 DAA34627.1 GBZX01002540 JAG90200.1 DS232258 EDS38575.1 CH933806 EDW16394.1
KPI98765.1 GDQN01004808 JAT86246.1 AGBW02008814 OWR52498.1 ODYU01011965 SOQ58010.1 KZ149914 PZC78022.1 NWSH01000174 PCG78753.1 PCG78754.1 JTDY01005451 KOB66987.1 KK852790 KDR16503.1 DS497681 EFA12034.1 GEZM01039645 JAV81300.1 GDAI01001561 JAI16042.1 AK417297 BAN20512.1 GFTR01002366 JAW14060.1 GDKW01001826 JAI54769.1 ACPB03017123 GAHY01001592 JAA75918.1 APGK01047102 BT126806 KB741077 KB632384 AEE61768.1 ENN74130.1 ERL94261.1 DS235869 EEB19432.1 GEDC01027381 JAS09917.1 GBHO01044545 GDHC01021505 JAF99058.1 JAP97123.1 KQ979608 KYN20508.1 GL888035 EGI68982.1 GBHO01044544 GDHC01001300 JAF99059.1 JAQ17329.1 GAKP01017745 JAC41207.1 GDHF01010935 GDHF01007070 GDHF01002034 JAI41379.1 JAI45244.1 JAI50280.1 GEBQ01001529 JAT38448.1 GBXI01013925 JAD00367.1 KK107144 QOIP01000003 EZA57609.1 RLU24319.1 JRES01000378 KNC31827.1 KQ976717 KYM76783.1 ADTU01023801 ADTU01023802 KQ414782 KOC61185.1 KQ977085 KYN05881.1 GFDG01001968 JAV16831.1 KQ434849 KZC08527.1 GECU01031045 GECU01022510 GECU01019250 JAS76661.1 JAS85196.1 JAS88456.1 GECZ01020513 JAS49256.1 GAMC01009590 JAB96965.1 GGMS01001898 MBY71101.1 LBMM01000401 KMQ98442.1 AXCM01000480 AXCN02001512 ADMH02002066 ETN59713.1 GANO01003442 JAB56429.1 JH431303 KA647882 AFP62511.1 GFXV01002873 MBW14678.1 KQ772519 OAD52447.1 NEVH01008204 PNF34749.1 KQ981650 KYN38645.1 KZ288224 PBC31941.1 JR050206 AEY61200.1 AAAB01008987 EAA01728.5 APCN01000828 ATLV01019123 KE525262 KFB43768.1 AXCP01002654 GALX01006770 JAB61696.1 GL764558 EFZ17457.1 ABLF02033604 CCAG010013859 JXJN01008014 CH477299 EAT44308.1 JXUM01033474 GAPW01005785 KQ560989 JAC07813.1 KXJ80110.1 GBRD01005056 JAG60765.1 GAMD01000710 JAB00881.1 GFPF01000763 MAA11909.1 GACK01006414 JAA58620.1 GEDV01009528 JAP79029.1 GEFH01005076 JAP63505.1 GFDL01012213 JAV22832.1 CH982788 EDX15763.1 CM000160 EDW98518.1 CH480828 EDW45232.1 CH954182 EDV53429.1 AE014297 AY084197 KX531576 AAF56692.1 AAL89935.1 ANY27386.1 GFAA01002056 JAU01379.1 BK007654 DAA34627.1 GBZX01002540 JAG90200.1 DS232258 EDS38575.1 CH933806 EDW16394.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000027135 UP000007266 UP000015103 UP000019118 UP000030742 UP000009046 UP000078492 UP000007755 UP000192223 UP000053097 UP000279307 UP000037069 UP000078540 UP000005205 UP000075881 UP000053825 UP000078542 UP000076502 UP000095300 UP000036403 UP000075883 UP000075885 UP000075886 UP000000673 UP000075920 UP000069272 UP000075900 UP000095301 UP000235965 UP000078541 UP000242457 UP000005203 UP000075902 UP000076407 UP000007062 UP000075840 UP000030765 UP000075880 UP000007819 UP000075903 UP000075882 UP000092444 UP000078200 UP000092445 UP000092460 UP000092443 UP000008820 UP000069940 UP000249989 UP000192221 UP000000304 UP000002282 UP000001292 UP000008711 UP000000803 UP000002358 UP000002320 UP000009192
UP000027135 UP000007266 UP000015103 UP000019118 UP000030742 UP000009046 UP000078492 UP000007755 UP000192223 UP000053097 UP000279307 UP000037069 UP000078540 UP000005205 UP000075881 UP000053825 UP000078542 UP000076502 UP000095300 UP000036403 UP000075883 UP000075885 UP000075886 UP000000673 UP000075920 UP000069272 UP000075900 UP000095301 UP000235965 UP000078541 UP000242457 UP000005203 UP000075902 UP000076407 UP000007062 UP000075840 UP000030765 UP000075880 UP000007819 UP000075903 UP000075882 UP000092444 UP000078200 UP000092445 UP000092460 UP000092443 UP000008820 UP000069940 UP000249989 UP000192221 UP000000304 UP000002282 UP000001292 UP000008711 UP000000803 UP000002358 UP000002320 UP000009192
Pfam
PF10224 DUF2205
Interpro
IPR019357
SCOC
ProteinModelPortal
H9JKZ6
S4PXK0
A0A3S2LTA3
A0A194Q5T8
A0A1E1WH64
A0A212FFJ1
+ More
A0A2H1WY60 A0A2W1BSG2 A0A2A4K4B8 A0A0L7KUN1 A0A067RCL3 D7EII5 A0A1Y1M9P2 A0A0K8TNS4 R4WRI2 A0A224XZK2 A0A0P4VS68 R4G849 J3JU93 E0W1C6 A0A1B6C8T3 A0A0A9VTK3 A0A195E5R6 F4W9Z2 A0A0A9W2F5 A0A1W4XAI2 A0A034VGK8 A0A0K8VR73 A0A1B6MRB1 A0A0A1WNH3 A0A026WRB0 A0A0L0CHZ0 A0A151HYK8 A0A158NR42 A0A182K7S3 A0A0L7QRC3 A0A151ILW5 A0A1L8EDN9 A0A154P9M7 A0A1B6HPT1 A0A1I8PFZ1 A0A1B6FGC6 W8B725 A0A2S2Q095 A0A0J7L642 A0A182ME14 A0A182PRQ8 A0A182QNT3 W5J8K2 A0A182VVW0 A0A182FRN3 A0A182RA37 U5EGH2 T1IQD3 T1PGF2 A0A2H8TN14 A0A310S530 A0A2J7R1N6 A0A151JWG4 A0A2A3ELN6 V9IJG7 A0A088AK63 A0A182TFC5 A0A182X9I2 Q7PX88 A0A182HKG5 A0A084W0M4 A0A182IK87 V5I7A6 E9IPP4 J9JLL5 A0A182URT8 A0A182L8P9 A0A1B0G6R5 A0A1A9UTP1 A0A1A9ZTB8 A0A1B0B3S5 A0A1A9YK27 Q17D62 A0A023EDY1 A0A1W4UR88 A0A0K8T6F9 T1DRA7 A0A224Y8N3 L7M3M5 A0A131YIE3 A0A131XBM5 A0A1Q3F5J6 B4NT73 B4PRS5 B4IC74 B3P629 Q9VB51 K7ISK8 A0A1E1XQ16 F0J9V3 A0A0C9QZM7 B0X1C6 B4K781
A0A2H1WY60 A0A2W1BSG2 A0A2A4K4B8 A0A0L7KUN1 A0A067RCL3 D7EII5 A0A1Y1M9P2 A0A0K8TNS4 R4WRI2 A0A224XZK2 A0A0P4VS68 R4G849 J3JU93 E0W1C6 A0A1B6C8T3 A0A0A9VTK3 A0A195E5R6 F4W9Z2 A0A0A9W2F5 A0A1W4XAI2 A0A034VGK8 A0A0K8VR73 A0A1B6MRB1 A0A0A1WNH3 A0A026WRB0 A0A0L0CHZ0 A0A151HYK8 A0A158NR42 A0A182K7S3 A0A0L7QRC3 A0A151ILW5 A0A1L8EDN9 A0A154P9M7 A0A1B6HPT1 A0A1I8PFZ1 A0A1B6FGC6 W8B725 A0A2S2Q095 A0A0J7L642 A0A182ME14 A0A182PRQ8 A0A182QNT3 W5J8K2 A0A182VVW0 A0A182FRN3 A0A182RA37 U5EGH2 T1IQD3 T1PGF2 A0A2H8TN14 A0A310S530 A0A2J7R1N6 A0A151JWG4 A0A2A3ELN6 V9IJG7 A0A088AK63 A0A182TFC5 A0A182X9I2 Q7PX88 A0A182HKG5 A0A084W0M4 A0A182IK87 V5I7A6 E9IPP4 J9JLL5 A0A182URT8 A0A182L8P9 A0A1B0G6R5 A0A1A9UTP1 A0A1A9ZTB8 A0A1B0B3S5 A0A1A9YK27 Q17D62 A0A023EDY1 A0A1W4UR88 A0A0K8T6F9 T1DRA7 A0A224Y8N3 L7M3M5 A0A131YIE3 A0A131XBM5 A0A1Q3F5J6 B4NT73 B4PRS5 B4IC74 B3P629 Q9VB51 K7ISK8 A0A1E1XQ16 F0J9V3 A0A0C9QZM7 B0X1C6 B4K781
PDB
4BWD
E-value=4.89646e-17,
Score=208
Ontologies
GO
PANTHER
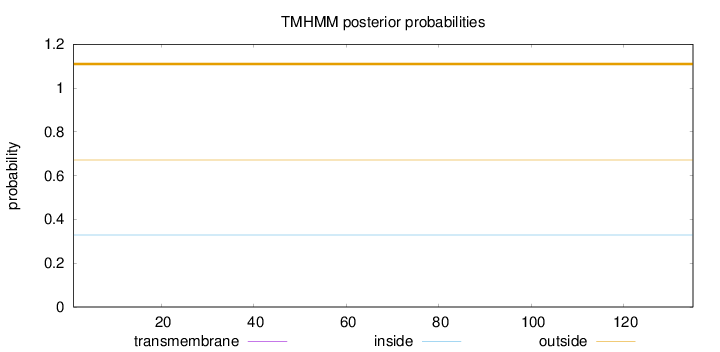
Topology
Length:
135
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00175
Exp number, first 60 AAs:
0
Total prob of N-in:
0.32913
outside
1 - 135
Population Genetic Test Statistics
Pi
280.555134
Theta
203.482736
Tajima's D
1.242236
CLR
0.012948
CSRT
0.72026398680066
Interpretation
Uncertain
