Gene
KWMTBOMO03865
Pre Gene Modal
BGIBMGA010174
Annotation
PREDICTED:_uncharacterized_protein_LOC101740250_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.131
Sequence
CDS
ATGTCTGCAAGTTTTGAACACACCGCTATGAAGAGCAGTTTCTCGCTGTCGTCACTGAGGAGCGCGCCCGCGCCGCCCGGCGGACCCTGCGAACAACTCGAGCGGCCTTACCATAGCCTCACTAAGAAGAGGAAGGAGCCGTGTAGAGAAGCCTGGCGCCGGAGTTGGGGCTCGACAGCGAGTGGCGGCAGCGCCGGCGACGATCTATGGCCGCTACTGCAACGACACTACGACTACATCATGGATAACCAGATCATCGACAGTTGCAAGGAAGCAAACGGCGAATTACTATCACATAATCCGTCAGCTTTGCTGTCTGGTCCGTGTCTACTGAACCCAACATCGGGGTTCAACAGACAGTGGTCGCTGAATCAGCTCGTATCCGAATTCGACGAGCTCTGCCAGTGGCTGACACACGTGCAGGAAGAAATATATTCTAGTCCCGAGAATTTGTCCAATTGGAAGCTTAGAGCGAATCGTATGGCAGAATTGGTGGCGGTCGAGCCGCGGCGTGCGAAGTTCGTTGAACAAGCGAATGCTATACTGGAGAGGATACCTGAAGCCAACGGAGAGGTGACCTGGCGTATTGAGCACTTGAGGATCAAGTGGGAAGCCCTTCGTCTGCTGCTCAGTCCGGAACAACCGCATCTTGACGGCGAGAATCCTGATACAGTTGACACCGCCCACGAGCTACGTTGCCTTCGTAGATGGTTGCACAGCATGGAAGGCCGCCTTCCACCACCCTCGCTTGCTGCAGCCCGCTCAGCCTCCCATCACGAGCTACTCAGGAAATTGAGAGAGCACCAGGTATGA
Protein
MSASFEHTAMKSSFSLSSLRSAPAPPGGPCEQLERPYHSLTKKRKEPCREAWRRSWGSTASGGSAGDDLWPLLQRHYDYIMDNQIIDSCKEANGELLSHNPSALLSGPCLLNPTSGFNRQWSLNQLVSEFDELCQWLTHVQEEIYSSPENLSNWKLRANRMAELVAVEPRRAKFVEQANAILERIPEANGEVTWRIEHLRIKWEALRLLLSPEQPHLDGENPDTVDTAHELRCLRRWLHSMEGRLPPPSLAAARSASHHELLRKLREHQV
Summary
Description
Tubulin is the major constituent of microtubules. It binds two moles of GTP, one at an exchangeable site on the beta chain and one at a non-exchangeable site on the alpha chain.
Subunit
Dimer of alpha and beta chains. A typical microtubule is a hollow water-filled tube with an outer diameter of 25 nm and an inner diameter of 15 nM. Alpha-beta heterodimers associate head-to-tail to form protofilaments running lengthwise along the microtubule wall with the beta-tubulin subunit facing the microtubule plus end conferring a structural polarity. Microtubules usually have 13 protofilaments but different protofilament numbers can be found in some organisms and specialized cells.
Similarity
Belongs to the tubulin family.
Uniprot
H9JKX3
A0A2A4K3B9
A0A3S2KWW8
A0A194RK33
A0A2W1BWM3
A0A194Q0E1
+ More
A0A154P087 A0A088AKL7 A0A0J7NPZ4 E2BRQ2 F4WDG2 A0A2A3ECI2 E9IZP9 A0A0N0BGY9 A0A2J7Q4T8 A0A0L7QQ29 A0A3L8DKT2 E2A9Q5 A0A026WM92 A0A158NIR1 A0A195BZ66 A0A151J7A1 A0A151X194 A0A195FRG0 K7JF83 A0A195CA67 A0A310SJM0 V5GRJ2 A0A1B6K0C3 A0A1B6DPW1 A0A1B6M7Q4 A0A1B6EWZ8 A0A1B6L1P0 A0A1B6LM59 A0A2R7WF41 A0A069DY63 A0A224XHT5 T1HCR3 A0A023EY65 T1IPR3 A0A0K8SH65 A0A0A9W446 A0A0K8SIL1 A0A0A9X443 A0A0A9WYK2
A0A154P087 A0A088AKL7 A0A0J7NPZ4 E2BRQ2 F4WDG2 A0A2A3ECI2 E9IZP9 A0A0N0BGY9 A0A2J7Q4T8 A0A0L7QQ29 A0A3L8DKT2 E2A9Q5 A0A026WM92 A0A158NIR1 A0A195BZ66 A0A151J7A1 A0A151X194 A0A195FRG0 K7JF83 A0A195CA67 A0A310SJM0 V5GRJ2 A0A1B6K0C3 A0A1B6DPW1 A0A1B6M7Q4 A0A1B6EWZ8 A0A1B6L1P0 A0A1B6LM59 A0A2R7WF41 A0A069DY63 A0A224XHT5 T1HCR3 A0A023EY65 T1IPR3 A0A0K8SH65 A0A0A9W446 A0A0K8SIL1 A0A0A9X443 A0A0A9WYK2
Pubmed
EMBL
BABH01011384
NWSH01000174
PCG78751.1
RSAL01002821
RVE40409.1
KQ460075
+ More
KPJ17907.1 KZ149914 PZC78024.1 KQ459582 KPI98768.1 KQ434787 KZC05326.1 LBMM01002656 KMQ94520.1 GL449978 EFN81646.1 GL888087 EGI67742.1 KZ288285 PBC29447.1 GL767232 EFZ13953.1 KQ435768 KOX75272.1 NEVH01018377 PNF23598.1 KQ414792 KOC60743.1 QOIP01000007 RLU21055.1 GL437918 EFN69857.1 KK107152 EZA57073.1 ADTU01017081 ADTU01017082 ADTU01017083 KQ976394 KYM93221.1 KQ979695 KYN19708.1 KQ982603 KYQ53961.1 KQ981285 KYN43180.1 AAZX01000619 AAZX01007219 AAZX01008461 AAZX01009647 AAZX01010443 KQ978068 KYM97585.1 KQ760551 OAD59858.1 GALX01005583 JAB62883.1 GECU01003103 JAT04604.1 GEDC01009590 JAS27708.1 GEBQ01008024 JAT31953.1 GECZ01027280 JAS42489.1 GEBQ01022350 JAT17627.1 GEBQ01015147 JAT24830.1 KK854735 PTY18287.1 GBGD01000202 JAC88687.1 GFTR01008825 JAW07601.1 ACPB03001290 ACPB03001291 GBBI01004710 JAC14002.1 JH431265 GBRD01013205 JAG52621.1 GBHO01043999 JAF99604.1 GBRD01013201 JAG52625.1 GBHO01030061 JAG13543.1 GBHO01030062 JAG13542.1
KPJ17907.1 KZ149914 PZC78024.1 KQ459582 KPI98768.1 KQ434787 KZC05326.1 LBMM01002656 KMQ94520.1 GL449978 EFN81646.1 GL888087 EGI67742.1 KZ288285 PBC29447.1 GL767232 EFZ13953.1 KQ435768 KOX75272.1 NEVH01018377 PNF23598.1 KQ414792 KOC60743.1 QOIP01000007 RLU21055.1 GL437918 EFN69857.1 KK107152 EZA57073.1 ADTU01017081 ADTU01017082 ADTU01017083 KQ976394 KYM93221.1 KQ979695 KYN19708.1 KQ982603 KYQ53961.1 KQ981285 KYN43180.1 AAZX01000619 AAZX01007219 AAZX01008461 AAZX01009647 AAZX01010443 KQ978068 KYM97585.1 KQ760551 OAD59858.1 GALX01005583 JAB62883.1 GECU01003103 JAT04604.1 GEDC01009590 JAS27708.1 GEBQ01008024 JAT31953.1 GECZ01027280 JAS42489.1 GEBQ01022350 JAT17627.1 GEBQ01015147 JAT24830.1 KK854735 PTY18287.1 GBGD01000202 JAC88687.1 GFTR01008825 JAW07601.1 ACPB03001290 ACPB03001291 GBBI01004710 JAC14002.1 JH431265 GBRD01013205 JAG52621.1 GBHO01043999 JAF99604.1 GBRD01013201 JAG52625.1 GBHO01030061 JAG13543.1 GBHO01030062 JAG13542.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000076502
+ More
UP000005203 UP000036403 UP000008237 UP000007755 UP000242457 UP000053105 UP000235965 UP000053825 UP000279307 UP000000311 UP000053097 UP000005205 UP000078540 UP000078492 UP000075809 UP000078541 UP000002358 UP000078542 UP000015103
UP000005203 UP000036403 UP000008237 UP000007755 UP000242457 UP000053105 UP000235965 UP000053825 UP000279307 UP000000311 UP000053097 UP000005205 UP000078540 UP000078492 UP000075809 UP000078541 UP000002358 UP000078542 UP000015103
PRIDE
Interpro
IPR002452
Alpha_tubulin
+ More
IPR017975 Tubulin_CS
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR037103 Tubulin/FtsZ_C_sf
IPR023123 Tubulin_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR012315 KASH
IPR039906 Anc-1-like
IPR039123 PPTC7
IPR036679 FlgN-like_sf
IPR018159 Spectrin/alpha-actinin
IPR017975 Tubulin_CS
IPR003008 Tubulin_FtsZ_GTPase
IPR008280 Tub_FtsZ_C
IPR037103 Tubulin/FtsZ_C_sf
IPR023123 Tubulin_C
IPR000217 Tubulin
IPR018316 Tubulin/FtsZ_2-layer-sand-dom
IPR036525 Tubulin/FtsZ_GTPase_sf
IPR012315 KASH
IPR039906 Anc-1-like
IPR039123 PPTC7
IPR036679 FlgN-like_sf
IPR018159 Spectrin/alpha-actinin
Gene 3D
ProteinModelPortal
H9JKX3
A0A2A4K3B9
A0A3S2KWW8
A0A194RK33
A0A2W1BWM3
A0A194Q0E1
+ More
A0A154P087 A0A088AKL7 A0A0J7NPZ4 E2BRQ2 F4WDG2 A0A2A3ECI2 E9IZP9 A0A0N0BGY9 A0A2J7Q4T8 A0A0L7QQ29 A0A3L8DKT2 E2A9Q5 A0A026WM92 A0A158NIR1 A0A195BZ66 A0A151J7A1 A0A151X194 A0A195FRG0 K7JF83 A0A195CA67 A0A310SJM0 V5GRJ2 A0A1B6K0C3 A0A1B6DPW1 A0A1B6M7Q4 A0A1B6EWZ8 A0A1B6L1P0 A0A1B6LM59 A0A2R7WF41 A0A069DY63 A0A224XHT5 T1HCR3 A0A023EY65 T1IPR3 A0A0K8SH65 A0A0A9W446 A0A0K8SIL1 A0A0A9X443 A0A0A9WYK2
A0A154P087 A0A088AKL7 A0A0J7NPZ4 E2BRQ2 F4WDG2 A0A2A3ECI2 E9IZP9 A0A0N0BGY9 A0A2J7Q4T8 A0A0L7QQ29 A0A3L8DKT2 E2A9Q5 A0A026WM92 A0A158NIR1 A0A195BZ66 A0A151J7A1 A0A151X194 A0A195FRG0 K7JF83 A0A195CA67 A0A310SJM0 V5GRJ2 A0A1B6K0C3 A0A1B6DPW1 A0A1B6M7Q4 A0A1B6EWZ8 A0A1B6L1P0 A0A1B6LM59 A0A2R7WF41 A0A069DY63 A0A224XHT5 T1HCR3 A0A023EY65 T1IPR3 A0A0K8SH65 A0A0A9W446 A0A0K8SIL1 A0A0A9X443 A0A0A9WYK2
Ontologies
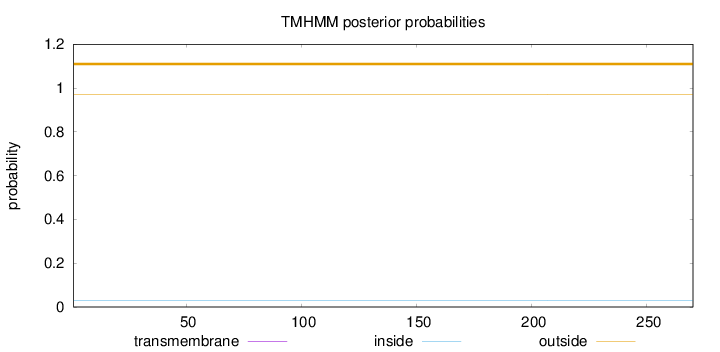
Topology
Subcellular location
Length:
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00022
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.03115
outside
1 - 270
Population Genetic Test Statistics
Pi
293.069475
Theta
170.492823
Tajima's D
2.265451
CLR
0.254372
CSRT
0.91770411479426
Interpretation
Uncertain
