Pre Gene Modal
BGIBMGA010177
Annotation
PREDICTED:_uncharacterized_protein_LOC101740250_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.892
Sequence
CDS
ATGTCCAGCGCTGAACGCAAAAAAACATTACTCGACAGCCTAGAGAGATCGAGGAACAACACTAGTCTAGAGAGCAACGACGGAGTCCGGCGCCGGAAGCCATCGGACATGGAACGGAAGAAACACTCACCAGAATTCGACTTGCTCAACGTGCAGGCTCTCGGCCGCCGACGTCACAGCTCCAACATGACCAGCGACGAGGTGAACAGCTCGCTCGAGTACACCGAAGTGAATAACAGACCGGACATTCTGGACTCGCTGAACCGACGTCGCAGGACCGAGAAAGACGGGGAGGCGGAGTTGCAAAAGGTCGCGCTTTCTGAACTGAGGCGGCGATCCACAGAGAGCATTGATTCTGAGGACGAGACGTCGAGTCCCAAGAAGCACAGCCGAAGGACGTGGACGGACGAATCGGAGTCGGAGGCGGAGGAAGTGAGGTCCCTGGTGCGGCGCTCGAGCTCGGCGCTGGACGCCGCCGAGGCGCTGCTGGAGCGGCACGAGGCCTCGCCCGACCTGCTGCGCGCCTTCGACTACACGGAGGTCGTGACTCGTTGTCGCGAAAATATCAACTTACTGGAAGCGAGTTTGGCTGGTGGTTCCCTTTCTGCGACATTGCAGCGCGATGTTAGAGCGGTGTCGGCTCGTTGGTCGGCTCTGCGTGCAGCCGCCACTCGTCGTGGTGGAGCCCGGCGACTCCGCAGGGAAATGGCAGCGCTTCGGGAAACGCTCGACGATATCTGTGACCCTGCAGACTGCGCTCCCCAACCGCACACAAGAGCTCAACTACACAGGAGAATCGAAGAGTTAAAGGATCGCCTATCGCGTCTTTTAGAATGCAAAGTTTCAATGTTGAAATTGACGGTCTCTGTACGTCGGACTCTCGGCGAGATGGAGTCAGATGATAGCGGTCTCGCTACTGAATTGGCAGCGTTATTATCCGCATGGGATGATGCGCATCAACGGACTTCTAATGAACTTCTCTCCTTGGAAAAAGCAATATGTGCTTGGGCCGAATGGGAGGGTGCGCTCCGCGAACTTCAAGGGGAATTGAGGGGAGATCTAGCTACTTTGGAGGCATTGCGTGATCGTGGCACAATCGTTGGTGACCAAACCGAAGTCGCCACTCAGATCAAACAATTGGCCGCATCTCTTATAGAGAAAAAGAAGGGTAGTTCAACATGCGACTCCCTCTCGGACTCTGGTATATCTGACGGAGATAGTGAAGGTGTTGGCAGACGGGCGAGACGTCTGGGTGCTCTGAGAGAGCTGGCGCGTCGTTTGCAAGCGACTTTAGCTCCGAATTCCCCAGCACACAGGGCTATTGCCAAGCGTATGGAACAAACCGAAAACGAAGTACGCACCTTACAAGCGTCTTGTAAAGCGCTGGTTGAACAAAACATACCGGAACTTAAAATTGACGATGACATAGAAAATAATGTCGGCCCAGTAGCTGGCAGTAAAACTGGCTCGGGCGATCCCGACTACAGCCCTCGAGGTGGATGGGTGTGGCGTGTATTACGTTCATCTCTCCCCATCCAACTGTGCCTTGTCGCTTTATTGTTAGCCGCTTGGTTGGTTGAACGTCCACGATGCTGCGATGCTCTTAACTCCCTTGCCCAATCGTTAACTCCTCAACTAAGATACGTTCGAGGACCTCCTCCAGTTTGA
Protein
MSSAERKKTLLDSLERSRNNTSLESNDGVRRRKPSDMERKKHSPEFDLLNVQALGRRRHSSNMTSDEVNSSLEYTEVNNRPDILDSLNRRRRTEKDGEAELQKVALSELRRRSTESIDSEDETSSPKKHSRRTWTDESESEAEEVRSLVRRSSSALDAAEALLERHEASPDLLRAFDYTEVVTRCRENINLLEASLAGGSLSATLQRDVRAVSARWSALRAAATRRGGARRLRREMAALRETLDDICDPADCAPQPHTRAQLHRRIEELKDRLSRLLECKVSMLKLTVSVRRTLGEMESDDSGLATELAALLSAWDDAHQRTSNELLSLEKAICAWAEWEGALRELQGELRGDLATLEALRDRGTIVGDQTEVATQIKQLAASLIEKKKGSSTCDSLSDSGISDGDSEGVGRRARRLGALRELARRLQATLAPNSPAHRAIAKRMEQTENEVRTLQASCKALVEQNIPELKIDDDIENNVGPVAGSKTGSGDPDYSPRGGWVWRVLRSSLPIQLCLVALLLAAWLVERPRCCDALNSLAQSLTPQLRYVRGPPPV
Summary
Uniprot
EMBL
Proteomes
Pfam
PF10541 KASH
ProteinModelPortal
Ontologies
PANTHER
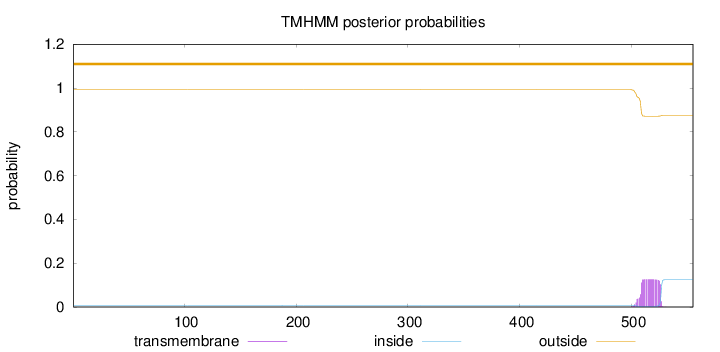
Topology
Length:
555
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.46095
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00652
outside
1 - 555
Population Genetic Test Statistics
Pi
281.370666
Theta
193.400335
Tajima's D
1.881392
CLR
0.109086
CSRT
0.857557122143893
Interpretation
Uncertain
