Gene
KWMTBOMO03852
Pre Gene Modal
BGIBMGA010182
Annotation
PREDICTED:_tubulin_polyglutamylase_TTLL4-like_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.245
Sequence
CDS
ATGAGCAAGGAGCAAAGGTTTCTGACTGCTTTGCATACGACAAATCCGTTTAAAACGATACCGTCACGCAAAGAATTGCCATTAATCATCCCAGAATCGATCTCAAGCTGTCTCCGACCGTCTCTCTTCCCGCGAGTGCCCCCATACCTGCGGTTCATCGGACACGAAGACTGCACTCCGCTGAAGGTTCCAGCTCCCATACAGAAGCATCTGAAATGGAAGCTCACAACGATAACGCCTTTAGTTGTGAAGAAAACTTTGACTAATTCTGGCTTTAGACTGGTTAAGAGCGAATGTGACACGTCAGAGTGTCCGCAAGAAGAGACCGTAGAGTGGATAGGCATATGGGGCAAACATATGAAGTCAATAATGTTCAGAGCCATAAAAGATGGACAGAAGATGAATCATTTCCCCGGTACCTTCCAAATCGGCAGGAAGGATCGATTGTGGAGGAACCTTCACAAACTTTGCACTAAGTATGGAGTCAAAGAATTTGGTATTATGCCGAAAACTTACGTATTGCCACATGATCTCAAAATGTTGAAGCATGACTGGGAAAAGTATGCCGCTAACAATGAAAGGTGGATCATAAAACCGCCTGCGTCAGCCCGAGGCACCGGCATCAAGGTGGTGTCCCGCTGGACGCAGATCCCGAAGAAGAGGCCGGTCGTCGTCCAGCGCTACGTCTCTAAACCGTATCTAATTAACGGAAGCAAGTTCGACATGAGGCTGTACGTGCTCGTCACGTCGGTGCACCCGCTCAGGATATATTTGTACAAAGATGGCCTTGCCAGGTTCGCGTCGGTAAAGTACAACGATGAGTTGACATCTCTGAACGATAGGTACATGCATTTGACTAATTACTCAATAAATAGGCTGTCCAAGAATTACACGCCAAACGAAGATTTTGCTTCTTGCGAGGGACACAAATGGACATTACAAACTCTATTCCAATACCTAAAGACTGAAAAGGACGTTGACACGGAAGCCCTGTGGGATTCCATGAAGGATCTCGTTATTAAAACCATAATATCGGGCGAGGCGAGCATCAGTTCGCTGACTAAAGCCAACATAACGTCGCGGTACAACTGCTACGAACTGTTCGGGATCGATGTTCTGCTGGACGAAGATCTCAAGCCCTGGCTCTTGGAGGTGAACATCTCGCCGAGCCTGCACAGCGCGTCGCCGCTCGACATCCACGTGAAGGGGCCCCTGGTCTCCACGGTGCTCAACATCGCGCAGTTCCAAGTGCCCAACAAAACCAACTTGGAAATGTTGGTTAAGGATAAACCACACCAATTAGCTGGATTACCATACGATGGCAGATTATACACAGTGTATTTATCCAAAGAAGAGAGAGACAAGCATATAATTTATACTAATATGGATGACAGAGATTTGTACCTGCGCGACATCCTGTCGACGCTGACGCCGGACGACGTGCGGCACCTGGTGCAGGCGGAGGACGAGCTGACGCAGGGCGGCGCCATGGAGCGCGTGTTCCCGACGCGCGACACGCACCGCTACCTGCCCTACCTCGCCGGCCCGCGATACTACAACAGACTCTTCGACGCCTGGGAGACCAGATACGGGCACAACAGGACCGCCGGCATCGAGCTGCTGCGCAACCTGTGCGACATCGGGTACCACCTGGAGGTGCCGCCCGTCCCGCTAAAGAATGACGTGGATGCGCCGTCGCCGCCCGTATCGGAGCGCCAGTCAGCGGGCTCCGTCTGCTCGGCGGGCGCGGGCCTGTCGCTCGCGGCGCCGCCCCCGCTCGAGGCGCGCGCCTAG
Protein
MSKEQRFLTALHTTNPFKTIPSRKELPLIIPESISSCLRPSLFPRVPPYLRFIGHEDCTPLKVPAPIQKHLKWKLTTITPLVVKKTLTNSGFRLVKSECDTSECPQEETVEWIGIWGKHMKSIMFRAIKDGQKMNHFPGTFQIGRKDRLWRNLHKLCTKYGVKEFGIMPKTYVLPHDLKMLKHDWEKYAANNERWIIKPPASARGTGIKVVSRWTQIPKKRPVVVQRYVSKPYLINGSKFDMRLYVLVTSVHPLRIYLYKDGLARFASVKYNDELTSLNDRYMHLTNYSINRLSKNYTPNEDFASCEGHKWTLQTLFQYLKTEKDVDTEALWDSMKDLVIKTIISGEASISSLTKANITSRYNCYELFGIDVLLDEDLKPWLLEVNISPSLHSASPLDIHVKGPLVSTVLNIAQFQVPNKTNLEMLVKDKPHQLAGLPYDGRLYTVYLSKEERDKHIIYTNMDDRDLYLRDILSTLTPDDVRHLVQAEDELTQGGAMERVFPTRDTHRYLPYLAGPRYYNRLFDAWETRYGHNRTAGIELLRNLCDIGYHLEVPPVPLKNDVDAPSPPVSERQSAGSVCSAGAGLSLAAPPPLEARA
Summary
Uniprot
H9JKY1
A0A2A4K4M1
A0A2H1VEH2
A0A194Q5V0
A0A194QTB7
A0A067R8J0
+ More
A0A2J7QZV7 A0A2J7QZW0 A0A2J7QZV6 A0A2J7QZW5 D6WYE8 A0A023EWR9 A0A182GJL8 A0A0T6B3W8 A0A1B6JU11 A0A1B6I987 A0A1B6JQF2 A0A1B6JXA3 A0A182WYN1 A0A023FBQ1 A0A1S4H082 A0A182I538 Q17JV3 A0A182KUI3 A0A084WC54 A0A182VAK4 A0A182NF19 A0A2M4BA23 A0A182MEP1 A0A182QQ02 A0A2M4BA66 A0A2M4BA54 A0A2M3Z0R9 A0A2M4A8B7 A0A2M4B9W0 A0A2M4CHA3 A0A2M4CHJ5 A0A2M3ZFV8 A0A2M4A7W0 A0A2M4A7L5 A0A2M4A7V4 A0A224XLX8 A0A182JZK4 A0A2M3ZG40 A0A182PFP2 A0A182UA81 Q7PVY2 A0A182WH41 A0A182INZ1 A0A182F2G2 E0VZA9 A0A182YCI6 A0A146MB87 W5JIV2 A0A1Q3F6A7 A0A1B6DER8 A0A1Q3F677 A0A1Q3F6B7 A0A182RIY8 A0A1B6C0P5 A0A1B6D6Y0 A0A1Q3F7C7 A0A1I8NC09 A0A2R7X3W7 A0A1I8NBY9 A0A1I8NBZ9 A0A1I8NBZ3 A0A336LVG9 A0A2H8TPL5 A0A0C9R429 A0A1Y1KUX1 A0A1W4VSP8 B0XFR1 A0A1S3CU55 B3N500 A0A1W4VST2 U5EQF5 A0A2S2P5Y8 A0A0Q5WAV4 B4P151 A0A1A9XV63 A0A1B0BTQ5 R4WTA1 A0A0J9R0J7 Q8IPB2 J9K688 A0A0P8XX36 A0A0R1DJQ7 A0A1S4E6M1 A0A1B0A5Y8 E9ICN0 A0A0P8ZGR4 Q8IGW4 Q9VKL9 A0A0J9R033 B3MJZ6 M9NEE5 A0A0L7QQY8 A0A0P8XGL7 A0A1B0FQW1
A0A2J7QZV7 A0A2J7QZW0 A0A2J7QZV6 A0A2J7QZW5 D6WYE8 A0A023EWR9 A0A182GJL8 A0A0T6B3W8 A0A1B6JU11 A0A1B6I987 A0A1B6JQF2 A0A1B6JXA3 A0A182WYN1 A0A023FBQ1 A0A1S4H082 A0A182I538 Q17JV3 A0A182KUI3 A0A084WC54 A0A182VAK4 A0A182NF19 A0A2M4BA23 A0A182MEP1 A0A182QQ02 A0A2M4BA66 A0A2M4BA54 A0A2M3Z0R9 A0A2M4A8B7 A0A2M4B9W0 A0A2M4CHA3 A0A2M4CHJ5 A0A2M3ZFV8 A0A2M4A7W0 A0A2M4A7L5 A0A2M4A7V4 A0A224XLX8 A0A182JZK4 A0A2M3ZG40 A0A182PFP2 A0A182UA81 Q7PVY2 A0A182WH41 A0A182INZ1 A0A182F2G2 E0VZA9 A0A182YCI6 A0A146MB87 W5JIV2 A0A1Q3F6A7 A0A1B6DER8 A0A1Q3F677 A0A1Q3F6B7 A0A182RIY8 A0A1B6C0P5 A0A1B6D6Y0 A0A1Q3F7C7 A0A1I8NC09 A0A2R7X3W7 A0A1I8NBY9 A0A1I8NBZ9 A0A1I8NBZ3 A0A336LVG9 A0A2H8TPL5 A0A0C9R429 A0A1Y1KUX1 A0A1W4VSP8 B0XFR1 A0A1S3CU55 B3N500 A0A1W4VST2 U5EQF5 A0A2S2P5Y8 A0A0Q5WAV4 B4P151 A0A1A9XV63 A0A1B0BTQ5 R4WTA1 A0A0J9R0J7 Q8IPB2 J9K688 A0A0P8XX36 A0A0R1DJQ7 A0A1S4E6M1 A0A1B0A5Y8 E9ICN0 A0A0P8ZGR4 Q8IGW4 Q9VKL9 A0A0J9R033 B3MJZ6 M9NEE5 A0A0L7QQY8 A0A0P8XGL7 A0A1B0FQW1
Pubmed
19121390
26354079
24845553
18362917
19820115
24945155
+ More
26483478 25474469 12364791 17510324 20966253 24438588 20566863 25244985 26823975 20920257 23761445 25315136 28004739 17994087 18057021 17550304 23691247 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665
26483478 25474469 12364791 17510324 20966253 24438588 20566863 25244985 26823975 20920257 23761445 25315136 28004739 17994087 18057021 17550304 23691247 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665
EMBL
BABH01011332
BABH01011333
NWSH01000134
PCG79205.1
ODYU01002131
SOQ39233.1
+ More
KQ459582 KPI98780.1 KQ461137 KPJ08768.1 KK852865 KDR14752.1 NEVH01009067 PNF34118.1 PNF34121.1 PNF34119.1 PNF34120.1 KQ971362 EFA09309.2 GAPW01000066 JAC13532.1 JXUM01068503 JXUM01068504 KQ562503 KXJ75780.1 LJIG01015987 KRT81953.1 GECU01005011 JAT02696.1 GECU01024218 JAS83488.1 GECU01006573 JAT01134.1 GECU01003872 JAT03835.1 GBBI01000359 JAC18353.1 AAAB01008984 APCN01003776 CH477230 EAT46959.1 ATLV01022565 KE525333 KFB47798.1 GGFJ01000742 MBW49883.1 AXCM01002215 AXCN02000831 GGFJ01000740 MBW49881.1 GGFJ01000741 MBW49882.1 GGFM01001362 MBW22113.1 GGFK01003670 MBW36991.1 GGFJ01000675 MBW49816.1 GGFL01000564 MBW64742.1 GGFL01000638 MBW64816.1 GGFM01006661 MBW27412.1 GGFK01003509 MBW36830.1 GGFK01003475 MBW36796.1 GGFK01003510 MBW36831.1 GFTR01007273 JAW09153.1 GGFM01006659 MBW27410.1 EAA14734.4 DS235851 EEB18715.1 GDHC01002124 JAQ16505.1 ADMH02001051 ETN64307.1 GFDL01011953 JAV23092.1 GEDC01013129 JAS24169.1 GFDL01012032 JAV23013.1 GFDL01011935 JAV23110.1 GEDC01030493 JAS06805.1 GEDC01015856 JAS21442.1 GFDL01011657 JAV23388.1 KK856858 PTY26488.1 UFQT01000225 SSX22042.1 GFXV01004318 MBW16123.1 GBYB01001565 GBYB01001566 JAG71332.1 JAG71333.1 GEZM01073290 JAV65182.1 DS232952 EDS26990.1 CH954177 EDV58945.2 KQS70583.1 KQS70585.1 GANO01004286 JAB55585.1 GGMR01012268 MBY24887.1 KQS70582.1 KQS70584.1 CM000157 EDW88026.2 KRJ97526.1 KRJ97529.1 JXJN01020305 AK417952 BAN21167.1 CM002910 KMY89668.1 KMY89670.1 KMY89671.1 AE014134 AAN10770.2 AAN10771.2 ACZ94234.1 ABLF02027168 ABLF02027174 CH902620 KPU73947.1 KPU73948.1 KRJ97527.1 KRJ97528.1 GL762326 EFZ21685.1 KPU73946.1 BT001555 AAN71310.1 AAF53045.2 ACZ94233.1 KMY89669.1 KMY89672.1 EDV32451.2 AFH03642.1 AFH03645.1 AFH03647.1 KQ414784 KOC61042.1 KPU73949.1 CCAG010010711
KQ459582 KPI98780.1 KQ461137 KPJ08768.1 KK852865 KDR14752.1 NEVH01009067 PNF34118.1 PNF34121.1 PNF34119.1 PNF34120.1 KQ971362 EFA09309.2 GAPW01000066 JAC13532.1 JXUM01068503 JXUM01068504 KQ562503 KXJ75780.1 LJIG01015987 KRT81953.1 GECU01005011 JAT02696.1 GECU01024218 JAS83488.1 GECU01006573 JAT01134.1 GECU01003872 JAT03835.1 GBBI01000359 JAC18353.1 AAAB01008984 APCN01003776 CH477230 EAT46959.1 ATLV01022565 KE525333 KFB47798.1 GGFJ01000742 MBW49883.1 AXCM01002215 AXCN02000831 GGFJ01000740 MBW49881.1 GGFJ01000741 MBW49882.1 GGFM01001362 MBW22113.1 GGFK01003670 MBW36991.1 GGFJ01000675 MBW49816.1 GGFL01000564 MBW64742.1 GGFL01000638 MBW64816.1 GGFM01006661 MBW27412.1 GGFK01003509 MBW36830.1 GGFK01003475 MBW36796.1 GGFK01003510 MBW36831.1 GFTR01007273 JAW09153.1 GGFM01006659 MBW27410.1 EAA14734.4 DS235851 EEB18715.1 GDHC01002124 JAQ16505.1 ADMH02001051 ETN64307.1 GFDL01011953 JAV23092.1 GEDC01013129 JAS24169.1 GFDL01012032 JAV23013.1 GFDL01011935 JAV23110.1 GEDC01030493 JAS06805.1 GEDC01015856 JAS21442.1 GFDL01011657 JAV23388.1 KK856858 PTY26488.1 UFQT01000225 SSX22042.1 GFXV01004318 MBW16123.1 GBYB01001565 GBYB01001566 JAG71332.1 JAG71333.1 GEZM01073290 JAV65182.1 DS232952 EDS26990.1 CH954177 EDV58945.2 KQS70583.1 KQS70585.1 GANO01004286 JAB55585.1 GGMR01012268 MBY24887.1 KQS70582.1 KQS70584.1 CM000157 EDW88026.2 KRJ97526.1 KRJ97529.1 JXJN01020305 AK417952 BAN21167.1 CM002910 KMY89668.1 KMY89670.1 KMY89671.1 AE014134 AAN10770.2 AAN10771.2 ACZ94234.1 ABLF02027168 ABLF02027174 CH902620 KPU73947.1 KPU73948.1 KRJ97527.1 KRJ97528.1 GL762326 EFZ21685.1 KPU73946.1 BT001555 AAN71310.1 AAF53045.2 ACZ94233.1 KMY89669.1 KMY89672.1 EDV32451.2 AFH03642.1 AFH03645.1 AFH03647.1 KQ414784 KOC61042.1 KPU73949.1 CCAG010010711
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000027135
UP000235965
+ More
UP000007266 UP000069940 UP000249989 UP000076407 UP000075840 UP000008820 UP000075882 UP000030765 UP000075903 UP000075884 UP000075883 UP000075886 UP000075881 UP000075885 UP000075902 UP000007062 UP000075920 UP000075880 UP000069272 UP000009046 UP000076408 UP000000673 UP000075900 UP000095301 UP000192221 UP000002320 UP000079169 UP000008711 UP000002282 UP000092443 UP000092460 UP000000803 UP000007819 UP000007801 UP000092445 UP000053825 UP000092444
UP000007266 UP000069940 UP000249989 UP000076407 UP000075840 UP000008820 UP000075882 UP000030765 UP000075903 UP000075884 UP000075883 UP000075886 UP000075881 UP000075885 UP000075902 UP000007062 UP000075920 UP000075880 UP000069272 UP000009046 UP000076408 UP000000673 UP000075900 UP000095301 UP000192221 UP000002320 UP000079169 UP000008711 UP000002282 UP000092443 UP000092460 UP000000803 UP000007819 UP000007801 UP000092445 UP000053825 UP000092444
Pfam
PF03133 TTL
Interpro
Gene 3D
ProteinModelPortal
H9JKY1
A0A2A4K4M1
A0A2H1VEH2
A0A194Q5V0
A0A194QTB7
A0A067R8J0
+ More
A0A2J7QZV7 A0A2J7QZW0 A0A2J7QZV6 A0A2J7QZW5 D6WYE8 A0A023EWR9 A0A182GJL8 A0A0T6B3W8 A0A1B6JU11 A0A1B6I987 A0A1B6JQF2 A0A1B6JXA3 A0A182WYN1 A0A023FBQ1 A0A1S4H082 A0A182I538 Q17JV3 A0A182KUI3 A0A084WC54 A0A182VAK4 A0A182NF19 A0A2M4BA23 A0A182MEP1 A0A182QQ02 A0A2M4BA66 A0A2M4BA54 A0A2M3Z0R9 A0A2M4A8B7 A0A2M4B9W0 A0A2M4CHA3 A0A2M4CHJ5 A0A2M3ZFV8 A0A2M4A7W0 A0A2M4A7L5 A0A2M4A7V4 A0A224XLX8 A0A182JZK4 A0A2M3ZG40 A0A182PFP2 A0A182UA81 Q7PVY2 A0A182WH41 A0A182INZ1 A0A182F2G2 E0VZA9 A0A182YCI6 A0A146MB87 W5JIV2 A0A1Q3F6A7 A0A1B6DER8 A0A1Q3F677 A0A1Q3F6B7 A0A182RIY8 A0A1B6C0P5 A0A1B6D6Y0 A0A1Q3F7C7 A0A1I8NC09 A0A2R7X3W7 A0A1I8NBY9 A0A1I8NBZ9 A0A1I8NBZ3 A0A336LVG9 A0A2H8TPL5 A0A0C9R429 A0A1Y1KUX1 A0A1W4VSP8 B0XFR1 A0A1S3CU55 B3N500 A0A1W4VST2 U5EQF5 A0A2S2P5Y8 A0A0Q5WAV4 B4P151 A0A1A9XV63 A0A1B0BTQ5 R4WTA1 A0A0J9R0J7 Q8IPB2 J9K688 A0A0P8XX36 A0A0R1DJQ7 A0A1S4E6M1 A0A1B0A5Y8 E9ICN0 A0A0P8ZGR4 Q8IGW4 Q9VKL9 A0A0J9R033 B3MJZ6 M9NEE5 A0A0L7QQY8 A0A0P8XGL7 A0A1B0FQW1
A0A2J7QZV7 A0A2J7QZW0 A0A2J7QZV6 A0A2J7QZW5 D6WYE8 A0A023EWR9 A0A182GJL8 A0A0T6B3W8 A0A1B6JU11 A0A1B6I987 A0A1B6JQF2 A0A1B6JXA3 A0A182WYN1 A0A023FBQ1 A0A1S4H082 A0A182I538 Q17JV3 A0A182KUI3 A0A084WC54 A0A182VAK4 A0A182NF19 A0A2M4BA23 A0A182MEP1 A0A182QQ02 A0A2M4BA66 A0A2M4BA54 A0A2M3Z0R9 A0A2M4A8B7 A0A2M4B9W0 A0A2M4CHA3 A0A2M4CHJ5 A0A2M3ZFV8 A0A2M4A7W0 A0A2M4A7L5 A0A2M4A7V4 A0A224XLX8 A0A182JZK4 A0A2M3ZG40 A0A182PFP2 A0A182UA81 Q7PVY2 A0A182WH41 A0A182INZ1 A0A182F2G2 E0VZA9 A0A182YCI6 A0A146MB87 W5JIV2 A0A1Q3F6A7 A0A1B6DER8 A0A1Q3F677 A0A1Q3F6B7 A0A182RIY8 A0A1B6C0P5 A0A1B6D6Y0 A0A1Q3F7C7 A0A1I8NC09 A0A2R7X3W7 A0A1I8NBY9 A0A1I8NBZ9 A0A1I8NBZ3 A0A336LVG9 A0A2H8TPL5 A0A0C9R429 A0A1Y1KUX1 A0A1W4VSP8 B0XFR1 A0A1S3CU55 B3N500 A0A1W4VST2 U5EQF5 A0A2S2P5Y8 A0A0Q5WAV4 B4P151 A0A1A9XV63 A0A1B0BTQ5 R4WTA1 A0A0J9R0J7 Q8IPB2 J9K688 A0A0P8XX36 A0A0R1DJQ7 A0A1S4E6M1 A0A1B0A5Y8 E9ICN0 A0A0P8ZGR4 Q8IGW4 Q9VKL9 A0A0J9R033 B3MJZ6 M9NEE5 A0A0L7QQY8 A0A0P8XGL7 A0A1B0FQW1
PDB
4YLS
E-value=4.94084e-44,
Score=450
Ontologies
GO
PANTHER
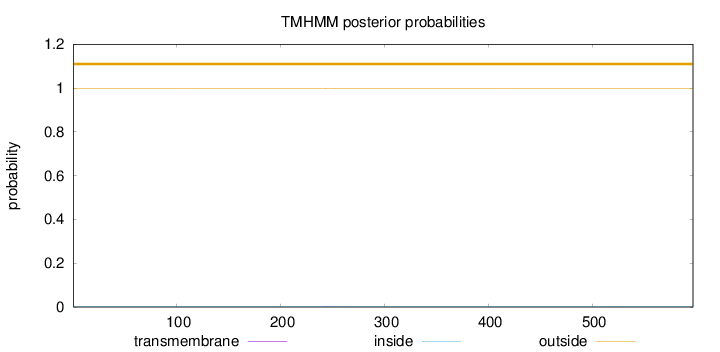
Topology
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00697
Exp number, first 60 AAs:
0.00055
Total prob of N-in:
0.00128
outside
1 - 597
Population Genetic Test Statistics
Pi
269.489125
Theta
177.529887
Tajima's D
1.129385
CLR
0.521686
CSRT
0.701814909254537
Interpretation
Uncertain
