Pre Gene Modal
BGIBMGA010193
Annotation
Short-chain_dehydrogenease/reductase_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.397 Mitochondrial Reliability : 1.393
Sequence
CDS
ATGGAATTTGGCGAAAAAGTTGTGTTGATAACCGGTGCAAGTTCGGGCATAGGTGCGGCCACAGCTATTCACTTTTCGAAATTATCGGCACAATTAGTACTTGTCGGTCGAAAAGAAAATAATTTACGTCGAATCGCGTTATACTGTGAAAAATCTAAAGGAATCAAACCTTTCACTATCACTGCTGATGTCACCGATGACGGCGACACCCAAAGAATAATTAATGAAACCATTGAACATTATGGAAAACTTGACGTTTTGATAAACAACGCCGGAGTGATCGGTATGGGCGGCATCAAAAATTCCACAATGGAAACTTATGACGACGTTATGTCGACTAATTTGAGGTCGGTTTATCGATTAACAATGCTTGCGGTTCCGCATCTAATACAAACTAAAGGATGTATCGTGAATTTGTCGTGTATAGCAGGTACAAAGCCCAGTACAATGGCTTTAGCCTATAACATCTCGAAAGCCGCATTAGATCACTTTACTAAATGTGTTGCGTTAGAATTAGCACCAGATGGCATTAGAGTGAATTCTGTTAACCCGGGATTTGTAAAAACGAACTTATTGAAAGGAATCGGATTAACTGAAGATCAATTAGAGTTGTTCGTAAAAAACGTTGTAGGGAACATGCCTCTAAAAAAGCCTGTTGAAAGCGATGAAGTTGCTGCGTTAATCGCCTTCCTTTCCAGTGATAAAGCGAAAAGTATTACAGGATCTATTTATGTTATCGATGGGGGCAGTTTGCTGCGCTGA
Protein
MEFGEKVVLITGASSGIGAATAIHFSKLSAQLVLVGRKENNLRRIALYCEKSKGIKPFTITADVTDDGDTQRIINETIEHYGKLDVLINNAGVIGMGGIKNSTMETYDDVMSTNLRSVYRLTMLAVPHLIQTKGCIVNLSCIAGTKPSTMALAYNISKAALDHFTKCVALELAPDGIRVNSVNPGFVKTNLLKGIGLTEDQLELFVKNVVGNMPLKKPVESDEVAALIAFLSSDKAKSITGSIYVIDGGSLLR
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JKZ2
A0A2H1VFS6
A0A2A4K5A7
A0A0L7K599
A0A212F7Q6
A0A194QTH4
+ More
A0A2H8TRX4 A0A194PZN2 J9K4P0 C4WUE4 A0A1S3IQP8 A0A2S2QKP0 F4WG13 A0A2H1V849 H9IVP2 A0A1S3IQ98 V3ZL06 A0A2A4JMG8 A0A2W1BPM7 Q2F652 A0A182TTS6 B0X121 A0A0L7KPN2 A0A067R623 A0A182V2E6 Q7QCA0 A0A0L7KZH2 A0A182M8T5 A0A1Q3FRU6 A0A158NTG0 A0A182LEW0 F4WG14 A0A2C9GR56 A0A194PHJ7 A0A1Q3FM45 A0A0J7KFU3 A0A084VXX9 A0A212FE39 A0A2J7QPM2 A0A151XBN1 A0A2J7QPM6 A0A182IKA3 A0A336MBZ2 A0A158NTG1 A0A336MQF8 A0A023ENR7 W5J801 A0A194RFH9 Q170J0 A0A194PR52 A0A182YHN0 A0A182Q2B3 A0A0L0CQ82 A0A182SS86 A0A1I8MT18 T1DQK7 J3JT59 A0A2M4BY41 A0A2M4BXH3 A0A1Q3FSC2 A0A1W7R596 Q16ZI8 A0A182FRI9 A0A2M4AJQ0 A0A2M3Z8L5 E0VS61 A0A2A4K610 A0A336LSD2 A0A158NTI2 S4PX81 I4DPW2 A0A2M4BX49 A0A1S4FHU5 A0A194RRD7 A0A182VPA1 A0A182TD91 A0A212F8M7 A0A1L8DJ46 A0A2M4BXA4 A0A182XHV8 Q7PR84 A0A182HSU0 A0A1J1IYY0 A0A2M4AJE9 A0A1L8DIV7 A0A0M9A8J7 A0A151JWN0 A0A182GNP8 I4DNP9 D6WBF1 A0A023ENP4 A0A182G1S0 A0A2P8Y4R7 A0A182IZ26 A0A182VWT1 A0A084VXY0 U5EY73 A0A2H1W6Q5 A0A182LEW9
A0A2H8TRX4 A0A194PZN2 J9K4P0 C4WUE4 A0A1S3IQP8 A0A2S2QKP0 F4WG13 A0A2H1V849 H9IVP2 A0A1S3IQ98 V3ZL06 A0A2A4JMG8 A0A2W1BPM7 Q2F652 A0A182TTS6 B0X121 A0A0L7KPN2 A0A067R623 A0A182V2E6 Q7QCA0 A0A0L7KZH2 A0A182M8T5 A0A1Q3FRU6 A0A158NTG0 A0A182LEW0 F4WG14 A0A2C9GR56 A0A194PHJ7 A0A1Q3FM45 A0A0J7KFU3 A0A084VXX9 A0A212FE39 A0A2J7QPM2 A0A151XBN1 A0A2J7QPM6 A0A182IKA3 A0A336MBZ2 A0A158NTG1 A0A336MQF8 A0A023ENR7 W5J801 A0A194RFH9 Q170J0 A0A194PR52 A0A182YHN0 A0A182Q2B3 A0A0L0CQ82 A0A182SS86 A0A1I8MT18 T1DQK7 J3JT59 A0A2M4BY41 A0A2M4BXH3 A0A1Q3FSC2 A0A1W7R596 Q16ZI8 A0A182FRI9 A0A2M4AJQ0 A0A2M3Z8L5 E0VS61 A0A2A4K610 A0A336LSD2 A0A158NTI2 S4PX81 I4DPW2 A0A2M4BX49 A0A1S4FHU5 A0A194RRD7 A0A182VPA1 A0A182TD91 A0A212F8M7 A0A1L8DJ46 A0A2M4BXA4 A0A182XHV8 Q7PR84 A0A182HSU0 A0A1J1IYY0 A0A2M4AJE9 A0A1L8DIV7 A0A0M9A8J7 A0A151JWN0 A0A182GNP8 I4DNP9 D6WBF1 A0A023ENP4 A0A182G1S0 A0A2P8Y4R7 A0A182IZ26 A0A182VWT1 A0A084VXY0 U5EY73 A0A2H1W6Q5 A0A182LEW9
Pubmed
EMBL
BABH01011332
ODYU01002131
SOQ39232.1
NWSH01000134
PCG79206.1
JTDY01010313
+ More
KOB58251.1 AGBW02009838 OWR49774.1 KQ461137 KPJ08767.1 GFXV01005148 MBW16953.1 KQ459582 KPI98781.1 ABLF02005808 AK341030 BAH71514.1 GGMS01008897 MBY78100.1 GL888128 EGI66780.1 ODYU01001173 SOQ37020.1 BABH01039662 KB203357 ESO84957.1 NWSH01000983 PCG73247.1 KZ149964 PZC76241.1 DQ311220 ABD36165.1 DS232250 EDS38413.1 JTDY01007623 KOB65070.1 KK852678 KDR18660.1 AAAB01008859 EAA07469.4 EGK96869.1 JTDY01004127 KOB68555.1 AXCM01010093 GFDL01004892 JAV30153.1 ADTU01025890 EGI66781.1 APCN01000350 KQ459604 KPI92523.1 GFDL01006355 JAV28690.1 LBMM01007925 KMQ89303.1 ATLV01018212 KE525224 KFB42823.1 AGBW02008979 OWR51994.1 NEVH01012088 PNF30536.1 KQ982320 KYQ57786.1 PNF30537.1 UFQT01000630 SSX25907.1 ADTU01025891 UFQS01001990 UFQT01001990 SSX12878.1 SSX32320.1 GAPW01003519 JAC10079.1 ADMH02002105 ETN58985.1 KQ460313 KPJ16055.1 EF173379 CH477473 ABM68625.1 EAT40351.1 KQ459595 KPI95931.1 AXCN02000007 JRES01000061 KNC34518.1 GAMD01001105 JAB00486.1 BT126415 KB632402 AEE61379.1 ERL94969.1 GGFJ01008517 MBW57658.1 GGFJ01008571 MBW57712.1 GFDL01004568 JAV30477.1 GEHC01001354 JAV46291.1 CH477489 EAT40086.1 GGFK01007692 MBW41013.1 GGFM01004116 MBW24867.1 DS235745 EEB16217.1 NWSH01000090 PCG79695.1 UFQS01000099 UFQT01000099 SSW99511.1 SSX19891.1 ADTU01025889 GAIX01005498 JAA87062.1 AK403809 BAM19952.1 GGFJ01008515 MBW57656.1 KQ459765 KPJ20037.1 AGBW02009702 OWR50087.1 GFDF01007697 JAV06387.1 GGFJ01008569 MBW57710.1 EAA07681.6 CVRI01000063 CRL04898.1 GGFK01007592 MBW40913.1 GFDF01007698 JAV06386.1 KQ435726 KOX78123.1 KQ981639 KYN38689.1 JXUM01076840 KQ562972 KXJ74733.1 AK403072 BAM19539.1 KQ971312 EEZ97890.1 GAPW01003549 JAC10049.1 JXUM01038912 KQ561184 KXJ79438.1 PYGN01000933 PSN39247.1 KFB42824.1 GANO01000479 JAB59392.1 ODYU01006626 SOQ48636.1
KOB58251.1 AGBW02009838 OWR49774.1 KQ461137 KPJ08767.1 GFXV01005148 MBW16953.1 KQ459582 KPI98781.1 ABLF02005808 AK341030 BAH71514.1 GGMS01008897 MBY78100.1 GL888128 EGI66780.1 ODYU01001173 SOQ37020.1 BABH01039662 KB203357 ESO84957.1 NWSH01000983 PCG73247.1 KZ149964 PZC76241.1 DQ311220 ABD36165.1 DS232250 EDS38413.1 JTDY01007623 KOB65070.1 KK852678 KDR18660.1 AAAB01008859 EAA07469.4 EGK96869.1 JTDY01004127 KOB68555.1 AXCM01010093 GFDL01004892 JAV30153.1 ADTU01025890 EGI66781.1 APCN01000350 KQ459604 KPI92523.1 GFDL01006355 JAV28690.1 LBMM01007925 KMQ89303.1 ATLV01018212 KE525224 KFB42823.1 AGBW02008979 OWR51994.1 NEVH01012088 PNF30536.1 KQ982320 KYQ57786.1 PNF30537.1 UFQT01000630 SSX25907.1 ADTU01025891 UFQS01001990 UFQT01001990 SSX12878.1 SSX32320.1 GAPW01003519 JAC10079.1 ADMH02002105 ETN58985.1 KQ460313 KPJ16055.1 EF173379 CH477473 ABM68625.1 EAT40351.1 KQ459595 KPI95931.1 AXCN02000007 JRES01000061 KNC34518.1 GAMD01001105 JAB00486.1 BT126415 KB632402 AEE61379.1 ERL94969.1 GGFJ01008517 MBW57658.1 GGFJ01008571 MBW57712.1 GFDL01004568 JAV30477.1 GEHC01001354 JAV46291.1 CH477489 EAT40086.1 GGFK01007692 MBW41013.1 GGFM01004116 MBW24867.1 DS235745 EEB16217.1 NWSH01000090 PCG79695.1 UFQS01000099 UFQT01000099 SSW99511.1 SSX19891.1 ADTU01025889 GAIX01005498 JAA87062.1 AK403809 BAM19952.1 GGFJ01008515 MBW57656.1 KQ459765 KPJ20037.1 AGBW02009702 OWR50087.1 GFDF01007697 JAV06387.1 GGFJ01008569 MBW57710.1 EAA07681.6 CVRI01000063 CRL04898.1 GGFK01007592 MBW40913.1 GFDF01007698 JAV06386.1 KQ435726 KOX78123.1 KQ981639 KYN38689.1 JXUM01076840 KQ562972 KXJ74733.1 AK403072 BAM19539.1 KQ971312 EEZ97890.1 GAPW01003549 JAC10049.1 JXUM01038912 KQ561184 KXJ79438.1 PYGN01000933 PSN39247.1 KFB42824.1 GANO01000479 JAB59392.1 ODYU01006626 SOQ48636.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000007819 UP000085678 UP000007755 UP000030746 UP000075902 UP000002320 UP000027135 UP000075903 UP000007062 UP000075883 UP000005205 UP000075882 UP000075840 UP000036403 UP000030765 UP000235965 UP000075809 UP000075880 UP000000673 UP000008820 UP000076408 UP000075886 UP000037069 UP000075901 UP000095301 UP000030742 UP000069272 UP000009046 UP000076407 UP000183832 UP000053105 UP000078541 UP000069940 UP000249989 UP000007266 UP000245037 UP000075920
UP000007819 UP000085678 UP000007755 UP000030746 UP000075902 UP000002320 UP000027135 UP000075903 UP000007062 UP000075883 UP000005205 UP000075882 UP000075840 UP000036403 UP000030765 UP000235965 UP000075809 UP000075880 UP000000673 UP000008820 UP000076408 UP000075886 UP000037069 UP000075901 UP000095301 UP000030742 UP000069272 UP000009046 UP000076407 UP000183832 UP000053105 UP000078541 UP000069940 UP000249989 UP000007266 UP000245037 UP000075920
Pfam
PF00106 adh_short
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JKZ2
A0A2H1VFS6
A0A2A4K5A7
A0A0L7K599
A0A212F7Q6
A0A194QTH4
+ More
A0A2H8TRX4 A0A194PZN2 J9K4P0 C4WUE4 A0A1S3IQP8 A0A2S2QKP0 F4WG13 A0A2H1V849 H9IVP2 A0A1S3IQ98 V3ZL06 A0A2A4JMG8 A0A2W1BPM7 Q2F652 A0A182TTS6 B0X121 A0A0L7KPN2 A0A067R623 A0A182V2E6 Q7QCA0 A0A0L7KZH2 A0A182M8T5 A0A1Q3FRU6 A0A158NTG0 A0A182LEW0 F4WG14 A0A2C9GR56 A0A194PHJ7 A0A1Q3FM45 A0A0J7KFU3 A0A084VXX9 A0A212FE39 A0A2J7QPM2 A0A151XBN1 A0A2J7QPM6 A0A182IKA3 A0A336MBZ2 A0A158NTG1 A0A336MQF8 A0A023ENR7 W5J801 A0A194RFH9 Q170J0 A0A194PR52 A0A182YHN0 A0A182Q2B3 A0A0L0CQ82 A0A182SS86 A0A1I8MT18 T1DQK7 J3JT59 A0A2M4BY41 A0A2M4BXH3 A0A1Q3FSC2 A0A1W7R596 Q16ZI8 A0A182FRI9 A0A2M4AJQ0 A0A2M3Z8L5 E0VS61 A0A2A4K610 A0A336LSD2 A0A158NTI2 S4PX81 I4DPW2 A0A2M4BX49 A0A1S4FHU5 A0A194RRD7 A0A182VPA1 A0A182TD91 A0A212F8M7 A0A1L8DJ46 A0A2M4BXA4 A0A182XHV8 Q7PR84 A0A182HSU0 A0A1J1IYY0 A0A2M4AJE9 A0A1L8DIV7 A0A0M9A8J7 A0A151JWN0 A0A182GNP8 I4DNP9 D6WBF1 A0A023ENP4 A0A182G1S0 A0A2P8Y4R7 A0A182IZ26 A0A182VWT1 A0A084VXY0 U5EY73 A0A2H1W6Q5 A0A182LEW9
A0A2H8TRX4 A0A194PZN2 J9K4P0 C4WUE4 A0A1S3IQP8 A0A2S2QKP0 F4WG13 A0A2H1V849 H9IVP2 A0A1S3IQ98 V3ZL06 A0A2A4JMG8 A0A2W1BPM7 Q2F652 A0A182TTS6 B0X121 A0A0L7KPN2 A0A067R623 A0A182V2E6 Q7QCA0 A0A0L7KZH2 A0A182M8T5 A0A1Q3FRU6 A0A158NTG0 A0A182LEW0 F4WG14 A0A2C9GR56 A0A194PHJ7 A0A1Q3FM45 A0A0J7KFU3 A0A084VXX9 A0A212FE39 A0A2J7QPM2 A0A151XBN1 A0A2J7QPM6 A0A182IKA3 A0A336MBZ2 A0A158NTG1 A0A336MQF8 A0A023ENR7 W5J801 A0A194RFH9 Q170J0 A0A194PR52 A0A182YHN0 A0A182Q2B3 A0A0L0CQ82 A0A182SS86 A0A1I8MT18 T1DQK7 J3JT59 A0A2M4BY41 A0A2M4BXH3 A0A1Q3FSC2 A0A1W7R596 Q16ZI8 A0A182FRI9 A0A2M4AJQ0 A0A2M3Z8L5 E0VS61 A0A2A4K610 A0A336LSD2 A0A158NTI2 S4PX81 I4DPW2 A0A2M4BX49 A0A1S4FHU5 A0A194RRD7 A0A182VPA1 A0A182TD91 A0A212F8M7 A0A1L8DJ46 A0A2M4BXA4 A0A182XHV8 Q7PR84 A0A182HSU0 A0A1J1IYY0 A0A2M4AJE9 A0A1L8DIV7 A0A0M9A8J7 A0A151JWN0 A0A182GNP8 I4DNP9 D6WBF1 A0A023ENP4 A0A182G1S0 A0A2P8Y4R7 A0A182IZ26 A0A182VWT1 A0A084VXY0 U5EY73 A0A2H1W6Q5 A0A182LEW9
PDB
1XHL
E-value=2.72637e-29,
Score=318
Ontologies
Topology
Subcellular location
Nucleus
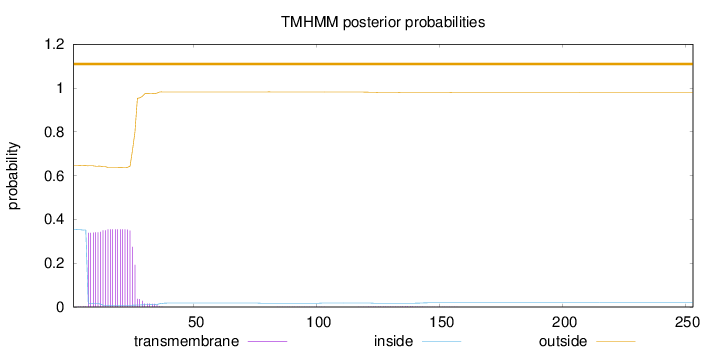
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.01591
Exp number, first 60 AAs:
6.95207
Total prob of N-in:
0.35402
outside
1 - 253
Population Genetic Test Statistics
Pi
25.618475
Theta
26.300741
Tajima's D
-0.07961
CLR
0.29162
CSRT
0.346282685865707
Interpretation
Uncertain
