Gene
KWMTBOMO03849
Pre Gene Modal
BGIBMGA010190
Annotation
PREDICTED:_uncharacterized_protein_LOC106116549_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.948
Sequence
CDS
ATGACTGTGCTGGAATCGTGTTGGTCTCCTTGCATTTGGTCCACTGATACAAAAATGGGAAGCAAGTTGGTAGCGGTTTATACTACCGCTATGAGCATAGTGCTGATTACGTTTATAGGATATCAAATGGACGGAGGCGATTCAACGCAGCTGTGGAACCCATTATTTGAAGCTGATGTTCGCGGATCCTTACAAATATTTGGAACAATATTTATAATAATATTGACACTGTTGATAATTTTTTCAATTTTAATGGTCATAGGCATCAACATATGGATGAGAGGTTTTATATTACCATGGCTCGCAACAATGGGATTCATCATAGTATTTCAATTCATTTTTGGTCTTTGGCTGCTGGGTGGATATTATATTTATTTGGACGCCACATTTGCAGCTCTGGCTGATTTCCTTTGGATGGCATATAATATATACTGTTGGCTGTGCGTGTTCTCTCAGTACCAAATAATATTGGAAATGCAATCTCCAAATATTGAAATCTTAGTTGAATATTAA
Protein
MTVLESCWSPCIWSTDTKMGSKLVAVYTTAMSIVLITFIGYQMDGGDSTQLWNPLFEADVRGSLQIFGTIFIIILTLLIIFSILMVIGINIWMRGFILPWLATMGFIIVFQFIFGLWLLGGYYIYLDATFAALADFLWMAYNIYCWLCVFSQYQIILEMQSPNIEILVEY
Summary
Uniprot
I4DQ15
A0A2A4JX05
A0A194RDB8
A0A2H1WPT7
A0A3S2NZU4
A0A1E1WLQ6
+ More
A0A2P8Z5T8 A0A1B6J3M5 A0A1B6C5B8 A0A1B6EHC1 A0A1L8D8Z5 A0A1W4XIM0 A0A1B6MNT8 U5EE31 A0A1I8PVM1 A0A224XZT3 A0A1B0G3L1 A0A1A9Z6E0 A0A1A9YDY5 A0A1A9V7X9 A0A023F8S4 A0A069DPU2 D6W7Z6 A0A0P4VU00 R4G915 K7IZK2 Q17J89 A0A1I8MKS4 A0A0V0GB16 A0A1Q3FXT2 A0A023EHW9 E2BVZ7 A0A0L0CIH2 A0A1Q3FXR8 A0A146LZD0 A0A026WNZ8 A0A0K8TN26 E0VX05 A0A3B0KIV5 A0A336LFN1 A0A182W146 Q294W5 B4GMI4 B4PKI7 A0A182NFZ4 B4QV41 B4IBC9 A0A084WNN7 A0A182FSU7 W5J5K3 A0A158NJJ6 A0A1Y1KPB3 B3P0G0 Q9VEF6 A0A0P8YM36 A0A1W4V566 B3LXE4 A0A195EJY7 A0A182XX48 A0A2M4C1F3 A0A2M3ZKT9 A0A182WV03 Q7QFT2 A0A182I8M8 B4JUG6 K7IZK3 E2A558 B4NFL5 B4NRF4 B4LUI5 A0A182SU51 B4KKP4 N6TYB7 A0A182RRU6 A0A182IQT6 A0A0M9A938 A0A151WJ90 A0A2M4AVB3 A0A195BFM7 A0A182GKH1 A0A2M4AUY1 A0A088AE46 A0A195F254 A0A2A3EAC5 F4X3X1 A0A182P2P3 A0A154PBA5 A0A182QHD9 A0A182LAH9 A0A0L7QRC2 A0A2S2PWG5 J9K652 A0A2S2N6L1 A0A1S3D194 T1IEY4
A0A2P8Z5T8 A0A1B6J3M5 A0A1B6C5B8 A0A1B6EHC1 A0A1L8D8Z5 A0A1W4XIM0 A0A1B6MNT8 U5EE31 A0A1I8PVM1 A0A224XZT3 A0A1B0G3L1 A0A1A9Z6E0 A0A1A9YDY5 A0A1A9V7X9 A0A023F8S4 A0A069DPU2 D6W7Z6 A0A0P4VU00 R4G915 K7IZK2 Q17J89 A0A1I8MKS4 A0A0V0GB16 A0A1Q3FXT2 A0A023EHW9 E2BVZ7 A0A0L0CIH2 A0A1Q3FXR8 A0A146LZD0 A0A026WNZ8 A0A0K8TN26 E0VX05 A0A3B0KIV5 A0A336LFN1 A0A182W146 Q294W5 B4GMI4 B4PKI7 A0A182NFZ4 B4QV41 B4IBC9 A0A084WNN7 A0A182FSU7 W5J5K3 A0A158NJJ6 A0A1Y1KPB3 B3P0G0 Q9VEF6 A0A0P8YM36 A0A1W4V566 B3LXE4 A0A195EJY7 A0A182XX48 A0A2M4C1F3 A0A2M3ZKT9 A0A182WV03 Q7QFT2 A0A182I8M8 B4JUG6 K7IZK3 E2A558 B4NFL5 B4NRF4 B4LUI5 A0A182SU51 B4KKP4 N6TYB7 A0A182RRU6 A0A182IQT6 A0A0M9A938 A0A151WJ90 A0A2M4AVB3 A0A195BFM7 A0A182GKH1 A0A2M4AUY1 A0A088AE46 A0A195F254 A0A2A3EAC5 F4X3X1 A0A182P2P3 A0A154PBA5 A0A182QHD9 A0A182LAH9 A0A0L7QRC2 A0A2S2PWG5 J9K652 A0A2S2N6L1 A0A1S3D194 T1IEY4
Pubmed
22651552
26354079
29403074
25474469
26334808
18362917
+ More
19820115 27129103 20075255 17510324 25315136 24945155 20798317 26108605 26823975 24508170 30249741 26369729 20566863 15632085 17994087 17550304 24438588 20920257 23761445 21347285 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 12364791 14747013 17210077 23537049 26483478 21719571 20966253
19820115 27129103 20075255 17510324 25315136 24945155 20798317 26108605 26823975 24508170 30249741 26369729 20566863 15632085 17994087 17550304 24438588 20920257 23761445 21347285 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 12364791 14747013 17210077 23537049 26483478 21719571 20966253
EMBL
AK403884
KQ459582
BAM20005.1
KPI98787.1
NWSH01000494
PCG76033.1
+ More
KQ460597 KPJ13906.1 ODYU01010104 SOQ54972.1 RSAL01000013 RVE53356.1 GDQN01003197 JAT87857.1 PYGN01000183 PSN51858.1 GECU01013921 JAS93785.1 GEDC01028596 JAS08702.1 GECZ01032446 JAS37323.1 GFDF01011229 JAV02855.1 GEBQ01002404 JAT37573.1 GANO01004475 JAB55396.1 GFTR01002817 JAW13609.1 CCAG010009033 GBBI01000915 JAC17797.1 GBGD01003242 JAC85647.1 KQ971307 EFA11022.2 GDKW01001104 JAI55491.1 GAHY01000012 JAA77498.1 CH477234 EAT46717.1 GECL01001501 JAP04623.1 GFDL01002665 JAV32380.1 GAPW01005122 GEHC01000354 JAC08476.1 JAV47291.1 GL451079 EFN80145.1 JRES01000345 KNC32045.1 GFDL01002676 JAV32369.1 GDHC01006929 GDHC01000528 JAQ11700.1 JAQ18101.1 KK107139 QOIP01000003 EZA57663.1 RLU24427.1 GDAI01002273 JAI15330.1 DS235824 EEB17911.1 OUUW01000014 SPP88460.1 UFQS01003189 UFQT01003189 SSX15345.1 SSX34717.1 CM000070 EAL28847.1 CH479185 EDW38058.1 CM000160 EDW95826.1 CM000364 EDX12524.1 CH480827 EDW44687.1 ATLV01024643 KE525354 KFB51831.1 ADMH02002125 ETN58678.1 ADTU01017959 GEZM01084724 JAV60707.1 CH954181 EDV48675.1 AE014297 AY071466 AAF55468.1 AAL49088.1 AFH06476.1 CH902617 KPU79905.1 EDV42788.2 KQ978782 KYN28456.1 GGFJ01010009 MBW59150.1 GGFM01008403 MBW29154.1 AAAB01008844 EAA06003.2 APCN01003089 CH916374 EDV91136.1 GL436875 EFN71425.1 CH964251 EDW83082.1 CH970975 EDW86943.1 CH940649 EDW64171.1 CH933807 EDW12708.1 APGK01050133 KB741166 KB632257 ENN73376.1 ERL90780.1 KQ435717 KOX78996.1 KQ983045 KYQ47906.1 GGFK01011406 MBW44727.1 KQ976490 KYM83403.1 JXUM01013188 JXUM01013189 KQ560371 KXJ82742.1 GGFK01011263 MBW44584.1 KQ981856 KYN34461.1 KZ288321 PBC28242.1 GL888624 EGI58918.1 KQ434849 KZC08518.1 AXCN02000343 KQ414782 KOC61178.1 GGMS01000633 MBY69836.1 ABLF02037361 GGMR01000141 MBY12760.1 ACPB03001339
KQ460597 KPJ13906.1 ODYU01010104 SOQ54972.1 RSAL01000013 RVE53356.1 GDQN01003197 JAT87857.1 PYGN01000183 PSN51858.1 GECU01013921 JAS93785.1 GEDC01028596 JAS08702.1 GECZ01032446 JAS37323.1 GFDF01011229 JAV02855.1 GEBQ01002404 JAT37573.1 GANO01004475 JAB55396.1 GFTR01002817 JAW13609.1 CCAG010009033 GBBI01000915 JAC17797.1 GBGD01003242 JAC85647.1 KQ971307 EFA11022.2 GDKW01001104 JAI55491.1 GAHY01000012 JAA77498.1 CH477234 EAT46717.1 GECL01001501 JAP04623.1 GFDL01002665 JAV32380.1 GAPW01005122 GEHC01000354 JAC08476.1 JAV47291.1 GL451079 EFN80145.1 JRES01000345 KNC32045.1 GFDL01002676 JAV32369.1 GDHC01006929 GDHC01000528 JAQ11700.1 JAQ18101.1 KK107139 QOIP01000003 EZA57663.1 RLU24427.1 GDAI01002273 JAI15330.1 DS235824 EEB17911.1 OUUW01000014 SPP88460.1 UFQS01003189 UFQT01003189 SSX15345.1 SSX34717.1 CM000070 EAL28847.1 CH479185 EDW38058.1 CM000160 EDW95826.1 CM000364 EDX12524.1 CH480827 EDW44687.1 ATLV01024643 KE525354 KFB51831.1 ADMH02002125 ETN58678.1 ADTU01017959 GEZM01084724 JAV60707.1 CH954181 EDV48675.1 AE014297 AY071466 AAF55468.1 AAL49088.1 AFH06476.1 CH902617 KPU79905.1 EDV42788.2 KQ978782 KYN28456.1 GGFJ01010009 MBW59150.1 GGFM01008403 MBW29154.1 AAAB01008844 EAA06003.2 APCN01003089 CH916374 EDV91136.1 GL436875 EFN71425.1 CH964251 EDW83082.1 CH970975 EDW86943.1 CH940649 EDW64171.1 CH933807 EDW12708.1 APGK01050133 KB741166 KB632257 ENN73376.1 ERL90780.1 KQ435717 KOX78996.1 KQ983045 KYQ47906.1 GGFK01011406 MBW44727.1 KQ976490 KYM83403.1 JXUM01013188 JXUM01013189 KQ560371 KXJ82742.1 GGFK01011263 MBW44584.1 KQ981856 KYN34461.1 KZ288321 PBC28242.1 GL888624 EGI58918.1 KQ434849 KZC08518.1 AXCN02000343 KQ414782 KOC61178.1 GGMS01000633 MBY69836.1 ABLF02037361 GGMR01000141 MBY12760.1 ACPB03001339
Proteomes
UP000053268
UP000218220
UP000053240
UP000283053
UP000245037
UP000192223
+ More
UP000095300 UP000092444 UP000092445 UP000092443 UP000078200 UP000007266 UP000002358 UP000008820 UP000095301 UP000008237 UP000037069 UP000053097 UP000279307 UP000009046 UP000268350 UP000075920 UP000001819 UP000008744 UP000002282 UP000075884 UP000000304 UP000001292 UP000030765 UP000069272 UP000000673 UP000005205 UP000008711 UP000000803 UP000007801 UP000192221 UP000078492 UP000076408 UP000076407 UP000007062 UP000075840 UP000001070 UP000000311 UP000007798 UP000008792 UP000075901 UP000009192 UP000019118 UP000030742 UP000075900 UP000075880 UP000053105 UP000075809 UP000078540 UP000069940 UP000249989 UP000005203 UP000078541 UP000242457 UP000007755 UP000075885 UP000076502 UP000075886 UP000075882 UP000053825 UP000007819 UP000079169 UP000015103
UP000095300 UP000092444 UP000092445 UP000092443 UP000078200 UP000007266 UP000002358 UP000008820 UP000095301 UP000008237 UP000037069 UP000053097 UP000279307 UP000009046 UP000268350 UP000075920 UP000001819 UP000008744 UP000002282 UP000075884 UP000000304 UP000001292 UP000030765 UP000069272 UP000000673 UP000005205 UP000008711 UP000000803 UP000007801 UP000192221 UP000078492 UP000076408 UP000076407 UP000007062 UP000075840 UP000001070 UP000000311 UP000007798 UP000008792 UP000075901 UP000009192 UP000019118 UP000030742 UP000075900 UP000075880 UP000053105 UP000075809 UP000078540 UP000069940 UP000249989 UP000005203 UP000078541 UP000242457 UP000007755 UP000075885 UP000076502 UP000075886 UP000075882 UP000053825 UP000007819 UP000079169 UP000015103
PRIDE
Pfam
PF15860 DUF4728
Interpro
IPR031720
DUF4728
ProteinModelPortal
I4DQ15
A0A2A4JX05
A0A194RDB8
A0A2H1WPT7
A0A3S2NZU4
A0A1E1WLQ6
+ More
A0A2P8Z5T8 A0A1B6J3M5 A0A1B6C5B8 A0A1B6EHC1 A0A1L8D8Z5 A0A1W4XIM0 A0A1B6MNT8 U5EE31 A0A1I8PVM1 A0A224XZT3 A0A1B0G3L1 A0A1A9Z6E0 A0A1A9YDY5 A0A1A9V7X9 A0A023F8S4 A0A069DPU2 D6W7Z6 A0A0P4VU00 R4G915 K7IZK2 Q17J89 A0A1I8MKS4 A0A0V0GB16 A0A1Q3FXT2 A0A023EHW9 E2BVZ7 A0A0L0CIH2 A0A1Q3FXR8 A0A146LZD0 A0A026WNZ8 A0A0K8TN26 E0VX05 A0A3B0KIV5 A0A336LFN1 A0A182W146 Q294W5 B4GMI4 B4PKI7 A0A182NFZ4 B4QV41 B4IBC9 A0A084WNN7 A0A182FSU7 W5J5K3 A0A158NJJ6 A0A1Y1KPB3 B3P0G0 Q9VEF6 A0A0P8YM36 A0A1W4V566 B3LXE4 A0A195EJY7 A0A182XX48 A0A2M4C1F3 A0A2M3ZKT9 A0A182WV03 Q7QFT2 A0A182I8M8 B4JUG6 K7IZK3 E2A558 B4NFL5 B4NRF4 B4LUI5 A0A182SU51 B4KKP4 N6TYB7 A0A182RRU6 A0A182IQT6 A0A0M9A938 A0A151WJ90 A0A2M4AVB3 A0A195BFM7 A0A182GKH1 A0A2M4AUY1 A0A088AE46 A0A195F254 A0A2A3EAC5 F4X3X1 A0A182P2P3 A0A154PBA5 A0A182QHD9 A0A182LAH9 A0A0L7QRC2 A0A2S2PWG5 J9K652 A0A2S2N6L1 A0A1S3D194 T1IEY4
A0A2P8Z5T8 A0A1B6J3M5 A0A1B6C5B8 A0A1B6EHC1 A0A1L8D8Z5 A0A1W4XIM0 A0A1B6MNT8 U5EE31 A0A1I8PVM1 A0A224XZT3 A0A1B0G3L1 A0A1A9Z6E0 A0A1A9YDY5 A0A1A9V7X9 A0A023F8S4 A0A069DPU2 D6W7Z6 A0A0P4VU00 R4G915 K7IZK2 Q17J89 A0A1I8MKS4 A0A0V0GB16 A0A1Q3FXT2 A0A023EHW9 E2BVZ7 A0A0L0CIH2 A0A1Q3FXR8 A0A146LZD0 A0A026WNZ8 A0A0K8TN26 E0VX05 A0A3B0KIV5 A0A336LFN1 A0A182W146 Q294W5 B4GMI4 B4PKI7 A0A182NFZ4 B4QV41 B4IBC9 A0A084WNN7 A0A182FSU7 W5J5K3 A0A158NJJ6 A0A1Y1KPB3 B3P0G0 Q9VEF6 A0A0P8YM36 A0A1W4V566 B3LXE4 A0A195EJY7 A0A182XX48 A0A2M4C1F3 A0A2M3ZKT9 A0A182WV03 Q7QFT2 A0A182I8M8 B4JUG6 K7IZK3 E2A558 B4NFL5 B4NRF4 B4LUI5 A0A182SU51 B4KKP4 N6TYB7 A0A182RRU6 A0A182IQT6 A0A0M9A938 A0A151WJ90 A0A2M4AVB3 A0A195BFM7 A0A182GKH1 A0A2M4AUY1 A0A088AE46 A0A195F254 A0A2A3EAC5 F4X3X1 A0A182P2P3 A0A154PBA5 A0A182QHD9 A0A182LAH9 A0A0L7QRC2 A0A2S2PWG5 J9K652 A0A2S2N6L1 A0A1S3D194 T1IEY4
Ontologies
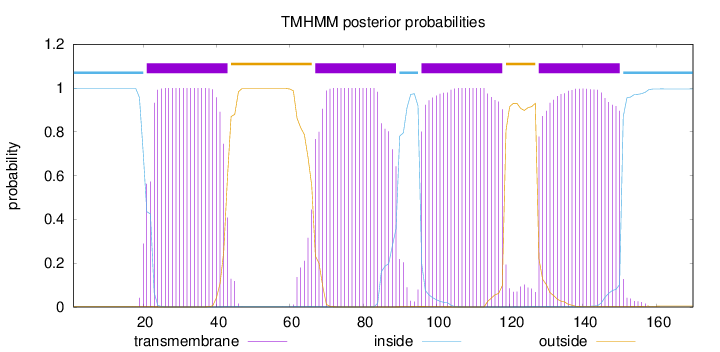
Topology
Length:
170
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
90.11041
Exp number, first 60 AAs:
21.64612
Total prob of N-in:
0.99785
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 66
TMhelix
67 - 89
inside
90 - 95
TMhelix
96 - 118
outside
119 - 127
TMhelix
128 - 150
inside
151 - 170
Population Genetic Test Statistics
Pi
259.608848
Theta
199.911998
Tajima's D
0.951293
CLR
0
CSRT
0.648967551622419
Interpretation
Uncertain
