Gene
KWMTBOMO03844
Annotation
PREDICTED:_uncharacterized_protein_LOC106710892_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 4.02
Sequence
CDS
ATGCTAGCCCTAAGCCCACTACCGTCACATTTGATAGAAAACAGTCAGAAGTACGAAATTAACAAAATTGAGACGGAAACTGACATAGATAAAGAACTTTGTGAAAATTTAAAGAAAATTCGTACCAGCCACATGAACGAGGAAGAAAAACGGGAAATTACTAAAATCTGCTATCAGTACCGTGACATATTCTACTCGGAAAACATTCCTTTATCGTTTACCCATACAGTAAAACACGAATTAAGACTAACCGACGACACCCCCATCTTTGTACGAAGTTATAGACAGGCTCCCCAACAACGAACAGAGATACAGAAACAGGTAGATAGTCTGTTAAAACAAGGAATCATTAGGGAAAGTATCTCCCCTTGGTCGTGCCCGGTACACATTGTTCCGAAAAAACCGGATGCATCAGGAAAAGTTAAATGGAGACTTGTTATTGACTATAGAAGACTTAATGACAGAATTATAGAAGACAAGTACCCCTTACCAAACATTAACGACATCCTTGACAGATTAGGGCGCGCACAATATTTCACGACCATAGATTTAGCAAGCGGCTACCATCAATTAGAAATGCACCCTAAAGACGTAGAGAAAACAGCGTTTACTACTGAAAGAGGCCACTATGAGTTCCTAAGAATGCCTTTCGGACTAAAAAATGCCCCGAGCACTTTCCAGCGTCTTATGGACCATATACTCCGAGGTATAGACAACGTATTTATGTACTTAGATTACGTCATAATAGCCGCGACGTCCCTACAAAACCACAATGAAAAACTGAAATTAGTATTTCAGCGATTCAAAATGCATAATCTGAAAGTTCAGTTAGACAAATCAGAATTTCTACAGAAGCACGTTAACTTTCTAGGACATGAATTGACTGACCAAGGACTAAATCCTAACAAGGACAAAATTAAAGCAGTATTAAATTTCCCTATACCACAAACGCAAAAAGACATAAAAGCTTTCTTAGGCCTAGTCGGGTACTATAGGAAGTTTATTAAGGACTTCGCGAAGTTGACGAAACCTTTAACAGCATGTCTAAAAAAGAACGCAAAGGTTGAACATACAAACGAATTTTTAGACGCAGTTGATAAATGCAAACAAATTCTAACAAACGCCCCAATCCTGCAATACCCTGACTTCGACAAACCGTTTATTTTAACGACAGACGCATCTGACTTTGCTTTAGGAGCAGTACTTTCCCAAGGCAATGTGGGCTCAGATAAACCAGTAGCCTATGCCTCAAGGACATTATCAGATACTGAGATCCGTTACTCTACCATAGAGAAAGAACTGTTAGGGATAGTATGGGCAATTAAGTATTTTAGACCTTACTTATATGGCCGTAAATTTACAATTTATACGGACCATAGACCCCTTACATGGTTAATGAGTCTAAAGGACCCTAACTCTAAATTAACACGATGGAAACTAAAGTTAGCAGAGTATGATTACAAAGTTGTTTATAAAAAGGGCAAACAAAACACTAACGCAGATGCACTATCTCGAGCAAAAATTTTTCATAATAGTATAGATTCTCTAGCTGTTAATGTTGATGACAATAGTGACGACAACATAATAAATAGAATATTCGAAAACGCCCGTAGACAGGCAGAGGTAGAAACTGATGACCCGAATAACCAAGACAATGAACGTAACAATACTGACAATGACGACCAACCTTATAACAACAATGACGTAGAAATGACCAGTATCTATCCCTCTCAAATTGACGAAGAGACAGACACAAGGAGTGCAACAACAGTCGACCCCGATCAATTAGTTTCTACAAATCATACCCAACCTGATAATGAAAATAACGGTATCCCTATAATATCCGACGCTATTGATAGACAGTTGAAACAATTTTACGTTAGGTCCACACCAGGTTCTACATACAGAGTAGAGGACAGATCAACAAACTCTAGGACAGTTATTAAGGATGTTTTCATCCCAGTAAATAACACTGAATCAGAAATTATCAAATTTTTAAAGGAACACACAATAGCTGACCGTGTTTTTCATTGCTATTTTTACGACGAAAATCTATACTTAGCCTTTTCAAGAGTGTATACTACGATATTTAATGACAGAGGACCTAAATTAATAAGATGTACTTCGCGGGTCACACTTGTTGAAAATAAAACTGAACAACAAGAACTCATTAAGCGATATCACGAAGGTAAATCATCGCATCGCGGTATCCAAGAGACCTTTAAGCATCTGCATAGGAATTATCACTGGCCTAATATGTTATTGACAGTTCAAAGGTTCATTAATCAATGCGACCTTTGCCTAAAGGCCAAATATGAAAGAAATCCTTTAAAACCTCCATTGATTATAACAGAGACACCTACGAAGCCATTTCAACACTTGTTCATGGATCTCTATAGTACTGGAGGTGCAACATTTTTAACAATTATCGACAATTTCTCTAAATTTGCCCAGGCGGTGCCTCTGAATGCTTCTAGTAGTGTTCACATCGCAGAAGCTCTATTACAAGTATTTTCTGTACTAGGACTACCTCTTAAAATCACCACAGACTCAGATACAAAGTTCGATAATGACGTCATAAAAGAGATATGTGCTTCGCATGATATCCACATTCACTTCACGACGCCTTACAACCCAAACTCTAACTCACCCATTGAACGATTTCATTCAACCATCGCAGAAATAATAAGAATTCAAAGAATGACAAATAAAGACGACCCCATACAATTGATCATGAAATACGCTATAATCGCCTATAACAACGCTATTCATTCTACTACAGGCTATACACCACGTGAGCTTTTATTCGGTCATACGGCATCCCGAAATCCATTAGAGCTATATTATCCTAAAGAATTTTATCAAGATTATGTCCTCAATCACCGCAAGAATGCAGAAGCAGTACAGGAATGTATAGCAGCCCACGTGTCTAAGAACAAAGAGCAGGTAATAGAAAAGAGAAACCAGGCAGCGCAAACAATCACGTTTAAGGTAGGTGAAACCGTTTACAAACAGGTCGCCAAAACCACCAGGAGCGACAAGACAAAACCAGTATTTAAAGATAAACAAAACCTCACCATTATTCCTAAATCCAAATACCTAGCACTGGGAACCAACGAGTACTCATACCTGGAGGAAGATTGCAAAAAGATCACACAAGACGTCCAACTCTGCACATCGCTGAACACCCAACCTGTGGAGAACTCTGAAGACTGCATAGTAACTCTTATAAAACACGAGAGCACAAACTGCACCCGTGCCAGGATGAACCTGAAACAAGGCAAGATCCAGAGACTAGAAGACAACAAATGGCTTATCATCTTGAAAGACGAACAAATCCTGAAATCTCGCTGCGGAAGGAAATCTGACTATAAAAAGATGTCAGGAATATACATCGCCAGCATTACAAGCGATTGTCAAGTGGAAATATTCAACCGAACACTGAAGACAAACACGGACACTATTACAGCTGATGAAATCGTACCCATTCCCAGCGAAACCACTATTCTAGAAGGGAATATTCGCTATAACCTACAACTGAAAGATATATCTCTGGATAGCATCCACGAACTGATGGACCGGGTTGAAAACATTCAACAACCTGTCATCGACTGGCAGACTATGATGACTACCCCAAGTTGGTCAACACTGGGACTCTACCTCATTCTGATAGCAATAATCATCTGGAAGCTGTGGCAGTGGAGACAGCGACGACTACAATCAAAGAACGAGAGCCCCGAGAACACTAGCATCGAGGACGCTGCTGGAAGCTGCGGGACGCGCTTCTATCTTAAGGAGGGAGGAGTTAGGCAATCGCCCGATGCCCGTATTTGCTGA
Protein
MLALSPLPSHLIENSQKYEINKIETETDIDKELCENLKKIRTSHMNEEEKREITKICYQYRDIFYSENIPLSFTHTVKHELRLTDDTPIFVRSYRQAPQQRTEIQKQVDSLLKQGIIRESISPWSCPVHIVPKKPDASGKVKWRLVIDYRRLNDRIIEDKYPLPNINDILDRLGRAQYFTTIDLASGYHQLEMHPKDVEKTAFTTERGHYEFLRMPFGLKNAPSTFQRLMDHILRGIDNVFMYLDYVIIAATSLQNHNEKLKLVFQRFKMHNLKVQLDKSEFLQKHVNFLGHELTDQGLNPNKDKIKAVLNFPIPQTQKDIKAFLGLVGYYRKFIKDFAKLTKPLTACLKKNAKVEHTNEFLDAVDKCKQILTNAPILQYPDFDKPFILTTDASDFALGAVLSQGNVGSDKPVAYASRTLSDTEIRYSTIEKELLGIVWAIKYFRPYLYGRKFTIYTDHRPLTWLMSLKDPNSKLTRWKLKLAEYDYKVVYKKGKQNTNADALSRAKIFHNSIDSLAVNVDDNSDDNIINRIFENARRQAEVETDDPNNQDNERNNTDNDDQPYNNNDVEMTSIYPSQIDEETDTRSATTVDPDQLVSTNHTQPDNENNGIPIISDAIDRQLKQFYVRSTPGSTYRVEDRSTNSRTVIKDVFIPVNNTESEIIKFLKEHTIADRVFHCYFYDENLYLAFSRVYTTIFNDRGPKLIRCTSRVTLVENKTEQQELIKRYHEGKSSHRGIQETFKHLHRNYHWPNMLLTVQRFINQCDLCLKAKYERNPLKPPLIITETPTKPFQHLFMDLYSTGGATFLTIIDNFSKFAQAVPLNASSSVHIAEALLQVFSVLGLPLKITTDSDTKFDNDVIKEICASHDIHIHFTTPYNPNSNSPIERFHSTIAEIIRIQRMTNKDDPIQLIMKYAIIAYNNAIHSTTGYTPRELLFGHTASRNPLELYYPKEFYQDYVLNHRKNAEAVQECIAAHVSKNKEQVIEKRNQAAQTITFKVGETVYKQVAKTTRSDKTKPVFKDKQNLTIIPKSKYLALGTNEYSYLEEDCKKITQDVQLCTSLNTQPVENSEDCIVTLIKHESTNCTRARMNLKQGKIQRLEDNKWLIILKDEQILKSRCGRKSDYKKMSGIYIASITSDCQVEIFNRTLKTNTDTITADEIVPIPSETTILEGNIRYNLQLKDISLDSIHELMDRVENIQQPVIDWQTMMTTPSWSTLGLYLILIAIIIWKLWQWRQRRLQSKNESPENTSIEDAAGSCGTRFYLKEGGVRQSPDARIC
Summary
Uniprot
EMBL
GEDC01017847
GEDC01011974
JAS19451.1
JAS25324.1
GDHC01021754
JAP96874.1
+ More
GBHO01012506 JAG31098.1 GBRD01016627 JAG49199.1 M32662 AAA92249.1 GFTR01008505 JAW07921.1 GBBI01004049 JAC14663.1 GBBI01004048 JAC14664.1 GFTR01008492 JAW07934.1 GBBI01004051 JAC14661.1 GANO01002504 JAB57367.1 GBHO01018861 JAG24743.1 GANO01002503 JAB57368.1 AF222049 AAF36671.1 GDAI01000737 JAI16866.1
GBHO01012506 JAG31098.1 GBRD01016627 JAG49199.1 M32662 AAA92249.1 GFTR01008505 JAW07921.1 GBBI01004049 JAC14663.1 GBBI01004048 JAC14664.1 GFTR01008492 JAW07934.1 GBBI01004051 JAC14661.1 GANO01002504 JAB57367.1 GBHO01018861 JAG24743.1 GANO01002503 JAB57368.1 AF222049 AAF36671.1 GDAI01000737 JAI16866.1
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
PDB
4OL8
E-value=4.59604e-76,
Score=729
Ontologies
KEGG
GO
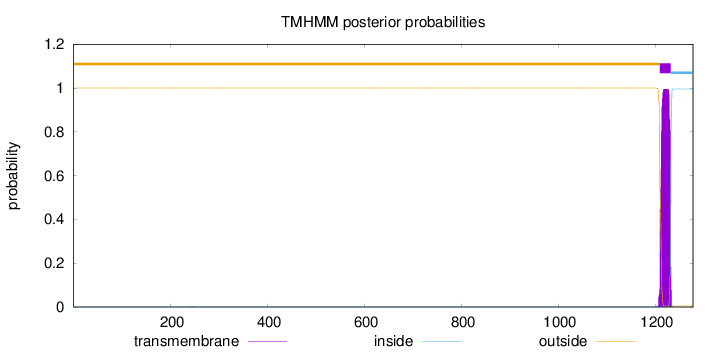
Topology
Length:
1278
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.18375
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00012
outside
1 - 1209
TMhelix
1210 - 1232
inside
1233 - 1278
Population Genetic Test Statistics
Pi
37.866634
Theta
37.444689
Tajima's D
-0.55784
CLR
355.311613
CSRT
0.232188390580471
Interpretation
Uncertain
