Gene
KWMTBOMO03836
Pre Gene Modal
BGIBMGA010002
Annotation
PREDICTED:_rho_GTPase-activating_protein_7_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.459
Sequence
CDS
ATGAAACTAGAGCACCACCATCATCAGAAGAGATCGAACGACTCTGACGATGAGCAATGCGCGCTGAGTGATAAATGGCAGTACGAGAGAAAGTCTAGAAGGTGGTCAAGAGTCGTCGAAATCACCCCACAAGCTCAAAGAAGATTACAGGTGATAGCAGCAAGAGCTCTAGCTGAGCGCGAAGCAAGAGAAGCCAGAGGTGAAGTGGAAGATGATGAAGACGAGACTCTGCCCACCATCAGGGTGGGACTGGTGGACGCAGATGGACATTACACTGACGTTGAGCCGGATCTTTTAAACATTCCTGGTCAACCAGTAATTCAGTTACCAAGCGACAAAGAAGATGAAGAAGAAACGGGTACCCGTTTCCGAAGGACCGGCTCGGAAAGACTACGCGATGGAGCCAAAGCATTGCTTCGTAGAGTTGAATCACTTAAGACTAGGAGAAGAAAACGCCAAAACAGAACCGAAGTTATCGTATCTTCGCCCCACTTTCTAGATGTACAAACCGACCAGTACCCAGATCTGAATTATATTGATATGACACCGACCTCGCCAACAGCATTCCCGTTCCCAGATTTCCATAGTTCACCAGCACACGCTGGTATCCGAACTCAGCCCCCTTCTCCTATGACCATAATGCCTCCATCACCGATAGCTCCTCTAGAACAATCCTTCGTCCTGAACCCTCCGTTCGGTGATGACAGTTCTTCATACGCGTCAGATGGCAGCATGAGTTCTAGTAACAAGAGCTCCAAGTCGAAACTCGGCCGAGCCAAAAGGATATTTAACAGAGGGCTCAAGAATGATGATTCAAGTGCTTTAAGTGATTCAGAATGTCAGCCGGCTAGTTGGAGACATAATTATTACAAAGAAATCAACACCCACAACACTGAGGTATATGTTGAACCACCATCACCGGTAGAGCTAAAAGCGGATCCTGAAATATTGCGGGAAGTACAAAAATCTCCAGGGCACACTCCACATAGGCGACAAGCAATAAGGACGAGTTCATTAAACCTAGGAAAGGAAGGACAAAAATTCCGCGACAGAAGTCTGAAAAGGGACAAATCTGCATCAAGGAGCTCTGAGCTTGATAGCAGTCCAAATTCTGCGGGTTCCAGGTCTCATGATGGCGATCAAGACGATGACGATGATAGCCTCGATCTGAAGAGTAAAAAGAGCAGTATACAAAGATGGTATTCCTTCCGTACTGGTGCCCTAAATGGAGGACCGAAAGCCAACAATACTCTTCAGCCTGCACCGAATCAAAAAGACCCTGACTCATACTTATCGCGACCAATGGCTTCATTATCCTGCGGACAATTACATATCTTGAGGAAGCTGGCTCTTCTAAGATTAACCGCCTGCATGGAGAGATACTGCCCATCACATAAATCCGGCTGGAATTGGGAGCTACCGAAACTTATACGGAAAATCAAAACTCCTGATTATAAAGATAAAACAGTATTTGGTGTACCCCTAACTGTGTCACTGCAAAGGACTGGTCACGCACTCCCCAAACCGATCCAGTGTGCTTTGAACTGGCTGAAAACCAATGCATTAGATCAGATGGGCATATTCAGGAAGGCCGGAGTAAAATCAAGGATTGCGAAACTTCGTAGCGTTGTCGAAGCAGCCGGTGCAGCTAATGCCGCATTCGCTATAGAGAACATGAACACCATCGATCAGAACCAGTCAAGCTTGAACTTCGACGGCGCTCAGGCTCATGACGTTGCGGACATGGTTAAGCAATACTTCAGAGAACTACCCGATGCATTACTCACAAATAAACTCAGCGAAACTTTTATTGCTATTTTTCAACATGTCCCTGAACCTCTTCGTCCTGATGCAGTGCAATGTGCATTACTACTGCTACCCGAAGAGCACTTGGAGGCTTTGCATTCACTACTCATATTTCTTTCTGACGTAGCCGAACATTCCGCGGTGAATCAAATGACAGCATCGAATTTGGCTGTTTGTCTGGCACCAACGTTACTGAGGCTGCATCACGCTCCACCGCAGACTGGGAGCACTAAAGAAGGGAATTCTAACAGGATGGTGATAGAAAGCGTAGCGGATCAAAGGCAGATCAGCGAAAGCCGGGCAGCCCACGCTTGTTTACTACTATTGATACAGAAACACATGCAATTATTTCTGGCTCCAGCTGACATGCTGGCCAGATGCAAATTCAACTACTTCGAAGAGAGTGTGCCGGTGTCATTAGAAGAATTGGGAGCTGATTTCAATCAAGACTGGAAAGGTTTTCAGCAGGCCTGCATCAAAGCTTTAATCAAAGAAGCCAAAGAAAAAACGAGAGGATGGATGTCAGTTTCTGGTGCGGCGCCGCATGTGGAACTCTGCTGTAAGAAAGTGGGAGACGGTCATCCACTCAGATTGTGGAGGGCGACTACAGACGTGGAAGCCCCGCCACAGGAGGTAGTTCAAAGAATCCTCCGCGAGCGGCATATTTGGGATGATTCGCTGGTGAAGTGGAGGATTGCAGAAAAACTTGGTGCCAACGCCGAAGTGTTTCAATACGTATCGGCTTCCTGCATCAATCTTCCGCCGCGGGAGTACTGCGTTTTGCGGTCGTGGCAGCAAAGCAGTGGTGGGAGCGGCAGCGGGGGTGGGGGCGGGGGCGGGTGGTGCGCCGTGGCGGAGACGTCGGTGGCGCACGGCGCGGCGCCGGCGTGCGGCCCGCGCGGCGTGGTGCTCGCCTCCCGGTACCTCGCGCAGCCGGCCGGCAAGGGGCGCAGCCGCCTCGTGCACCTGGCCAGGGTCGACACCATGGGTCGAACGCCAGAATGGTACAATAAATGCTACGGGCACATCTGCGCCCTGTACTTGGCCAGGATTCGAGCCTCGTTCAAGCACAACACGGAAGGCCCGGAATCCAACGTATGA
Protein
MKLEHHHHQKRSNDSDDEQCALSDKWQYERKSRRWSRVVEITPQAQRRLQVIAARALAEREAREARGEVEDDEDETLPTIRVGLVDADGHYTDVEPDLLNIPGQPVIQLPSDKEDEEETGTRFRRTGSERLRDGAKALLRRVESLKTRRRKRQNRTEVIVSSPHFLDVQTDQYPDLNYIDMTPTSPTAFPFPDFHSSPAHAGIRTQPPSPMTIMPPSPIAPLEQSFVLNPPFGDDSSSYASDGSMSSSNKSSKSKLGRAKRIFNRGLKNDDSSALSDSECQPASWRHNYYKEINTHNTEVYVEPPSPVELKADPEILREVQKSPGHTPHRRQAIRTSSLNLGKEGQKFRDRSLKRDKSASRSSELDSSPNSAGSRSHDGDQDDDDDSLDLKSKKSSIQRWYSFRTGALNGGPKANNTLQPAPNQKDPDSYLSRPMASLSCGQLHILRKLALLRLTACMERYCPSHKSGWNWELPKLIRKIKTPDYKDKTVFGVPLTVSLQRTGHALPKPIQCALNWLKTNALDQMGIFRKAGVKSRIAKLRSVVEAAGAANAAFAIENMNTIDQNQSSLNFDGAQAHDVADMVKQYFRELPDALLTNKLSETFIAIFQHVPEPLRPDAVQCALLLLPEEHLEALHSLLIFLSDVAEHSAVNQMTASNLAVCLAPTLLRLHHAPPQTGSTKEGNSNRMVIESVADQRQISESRAAHACLLLLIQKHMQLFLAPADMLARCKFNYFEESVPVSLEELGADFNQDWKGFQQACIKALIKEAKEKTRGWMSVSGAAPHVELCCKKVGDGHPLRLWRATTDVEAPPQEVVQRILRERHIWDDSLVKWRIAEKLGANAEVFQYVSASCINLPPREYCVLRSWQQSSGGSGSGGGGGGGWCAVAETSVAHGAAPACGPRGVVLASRYLAQPAGKGRSRLVHLARVDTMGRTPEWYNKCYGHICALYLARIRASFKHNTEGPESNV
Summary
Uniprot
A0A2A4JG32
A0A2H1W8Z7
A0A212FJT3
A0A194PZP2
A0A194R9G0
A0A2A4JHI2
+ More
H9JKF1 E9I8Y7 A0A158NJR5 A0A195BFN7 A0A195C8F3 A0A195F243 A0A151WJA7 A0A3L8DVB6 F4X3X9 A0A2J7RTI3 A0A2J7RTI7 A0A026WPT9 A0A195EKP6 A0A0C9RRR6 K7IZK0 A0A310SPY5 A0A2A3EPY6 A0A088A7P6 A0A232FMY5 E0VX09 A0A154P9C2 A0A1L8DL91 A0A1L8DL04 A0A1B0CDI4 A0A1B6J9H9 A0A0J7L738 A0A1B6E836 A0A0C9R3I9 A0A1B6EBR3 A0A224XGN9 A0A0P4VHB2 A0A1L8DL93 Q297U8 B4HEN4 Q9VFK2 B3P0Q5 B4LY65 B4QZV4 Q8SY28 B4PST6 A0A1W4US81 B3LVG1 A0A3B0JIW8 B0W210 A0A182GUA4 A8WHF8 A0A0R3NHI5 A0A0Q9WNS7 A8JR05 A0A0R1E424 A0A1W4V4I9 A0A0N8P181 A0A0P8XWW0 A0A1J1HL51 A0A1Q3G475 A0A1Q3G469 Q17FX7 W8AV38 W8AR15 W8AJ15 W8BFR5 A0A0L0BKW0 A0A0K8V161 A0A0K8V6V7 A0A0A1XKZ3 A0A0K8UWK8 A0A0K8VI39 A0A0A1XEQ4 B4JTT0 A0A1I8QD82 W8AR19 A0A034V721
H9JKF1 E9I8Y7 A0A158NJR5 A0A195BFN7 A0A195C8F3 A0A195F243 A0A151WJA7 A0A3L8DVB6 F4X3X9 A0A2J7RTI3 A0A2J7RTI7 A0A026WPT9 A0A195EKP6 A0A0C9RRR6 K7IZK0 A0A310SPY5 A0A2A3EPY6 A0A088A7P6 A0A232FMY5 E0VX09 A0A154P9C2 A0A1L8DL91 A0A1L8DL04 A0A1B0CDI4 A0A1B6J9H9 A0A0J7L738 A0A1B6E836 A0A0C9R3I9 A0A1B6EBR3 A0A224XGN9 A0A0P4VHB2 A0A1L8DL93 Q297U8 B4HEN4 Q9VFK2 B3P0Q5 B4LY65 B4QZV4 Q8SY28 B4PST6 A0A1W4US81 B3LVG1 A0A3B0JIW8 B0W210 A0A182GUA4 A8WHF8 A0A0R3NHI5 A0A0Q9WNS7 A8JR05 A0A0R1E424 A0A1W4V4I9 A0A0N8P181 A0A0P8XWW0 A0A1J1HL51 A0A1Q3G475 A0A1Q3G469 Q17FX7 W8AV38 W8AR15 W8AJ15 W8BFR5 A0A0L0BKW0 A0A0K8V161 A0A0K8V6V7 A0A0A1XKZ3 A0A0K8UWK8 A0A0K8VI39 A0A0A1XEQ4 B4JTT0 A0A1I8QD82 W8AR19 A0A034V721
Pubmed
EMBL
NWSH01001501
PCG71045.1
ODYU01007102
SOQ49575.1
AGBW02008227
OWR53986.1
+ More
KQ459582 KPI98791.1 KQ460597 KPJ13900.1 PCG71044.1 BABH01024929 BABH01024930 GL761662 EFZ22994.1 ADTU01017962 ADTU01017963 ADTU01017964 ADTU01017965 ADTU01017966 ADTU01017967 ADTU01017968 ADTU01017969 ADTU01017970 ADTU01017971 KQ976490 KYM83413.1 KQ978081 KYM97107.1 KQ981856 KYN34451.1 KQ983045 KYQ47897.1 QOIP01000003 RLU24430.1 GL888624 EGI58926.1 NEVH01000001 PNF44155.1 PNF44152.1 KK107139 EZA57666.1 KQ978782 KYN28444.1 GBYB01010071 JAG79838.1 KQ761164 OAD58101.1 KZ288203 PBC33332.1 NNAY01000007 OXU32096.1 DS235824 EEB17915.1 KQ434849 KZC08509.1 GFDF01006947 JAV07137.1 GFDF01006948 JAV07136.1 AJWK01007887 GECU01011917 JAS95789.1 LBMM01000424 KMQ98393.1 GEDC01003215 JAS34083.1 GBYB01002515 JAG72282.1 GEDC01001922 JAS35376.1 GFTR01008674 JAW07752.1 GDKW01003614 JAI52981.1 GFDF01006872 JAV07212.1 CM000070 EAL28107.1 CH480815 EDW42191.1 AE014297 BT044146 AAF55053.2 ACH92211.1 CH954181 EDV48881.1 CH940650 EDW67953.1 CM000364 EDX12952.1 AY075440 AAL68254.1 CM000160 EDW97582.1 CH902617 EDV41489.1 OUUW01000005 SPP80272.1 DS231824 EDS27446.1 JXUM01089191 KQ563752 KXJ73370.1 BT031104 ABX00726.1 KRT00462.1 KRF83574.1 KRF83575.1 ABW08670.1 ABW08671.1 KRK03845.1 KPU79188.1 KPU79189.1 KPU79187.1 KPU79190.1 CVRI01000006 CRK88132.1 GFDL01000442 JAV34603.1 GFDL01000448 JAV34597.1 CH477267 EAT45497.2 GAMC01018057 JAB88498.1 GAMC01018058 JAB88497.1 GAMC01018055 JAB88500.1 GAMC01018056 JAB88499.1 JRES01001706 KNC20687.1 GDHF01019723 JAI32591.1 GDHF01017647 JAI34667.1 GBXI01002675 JAD11617.1 GDHF01033799 GDHF01021401 GDHF01012625 JAI18515.1 JAI30913.1 JAI39689.1 GDHF01013745 JAI38569.1 GBXI01004886 JAD09406.1 CH916374 EDV91509.1 GAMC01018053 JAB88502.1 GAKP01021402 JAC37550.1
KQ459582 KPI98791.1 KQ460597 KPJ13900.1 PCG71044.1 BABH01024929 BABH01024930 GL761662 EFZ22994.1 ADTU01017962 ADTU01017963 ADTU01017964 ADTU01017965 ADTU01017966 ADTU01017967 ADTU01017968 ADTU01017969 ADTU01017970 ADTU01017971 KQ976490 KYM83413.1 KQ978081 KYM97107.1 KQ981856 KYN34451.1 KQ983045 KYQ47897.1 QOIP01000003 RLU24430.1 GL888624 EGI58926.1 NEVH01000001 PNF44155.1 PNF44152.1 KK107139 EZA57666.1 KQ978782 KYN28444.1 GBYB01010071 JAG79838.1 KQ761164 OAD58101.1 KZ288203 PBC33332.1 NNAY01000007 OXU32096.1 DS235824 EEB17915.1 KQ434849 KZC08509.1 GFDF01006947 JAV07137.1 GFDF01006948 JAV07136.1 AJWK01007887 GECU01011917 JAS95789.1 LBMM01000424 KMQ98393.1 GEDC01003215 JAS34083.1 GBYB01002515 JAG72282.1 GEDC01001922 JAS35376.1 GFTR01008674 JAW07752.1 GDKW01003614 JAI52981.1 GFDF01006872 JAV07212.1 CM000070 EAL28107.1 CH480815 EDW42191.1 AE014297 BT044146 AAF55053.2 ACH92211.1 CH954181 EDV48881.1 CH940650 EDW67953.1 CM000364 EDX12952.1 AY075440 AAL68254.1 CM000160 EDW97582.1 CH902617 EDV41489.1 OUUW01000005 SPP80272.1 DS231824 EDS27446.1 JXUM01089191 KQ563752 KXJ73370.1 BT031104 ABX00726.1 KRT00462.1 KRF83574.1 KRF83575.1 ABW08670.1 ABW08671.1 KRK03845.1 KPU79188.1 KPU79189.1 KPU79187.1 KPU79190.1 CVRI01000006 CRK88132.1 GFDL01000442 JAV34603.1 GFDL01000448 JAV34597.1 CH477267 EAT45497.2 GAMC01018057 JAB88498.1 GAMC01018058 JAB88497.1 GAMC01018055 JAB88500.1 GAMC01018056 JAB88499.1 JRES01001706 KNC20687.1 GDHF01019723 JAI32591.1 GDHF01017647 JAI34667.1 GBXI01002675 JAD11617.1 GDHF01033799 GDHF01021401 GDHF01012625 JAI18515.1 JAI30913.1 JAI39689.1 GDHF01013745 JAI38569.1 GBXI01004886 JAD09406.1 CH916374 EDV91509.1 GAMC01018053 JAB88502.1 GAKP01021402 JAC37550.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
UP000005205
+ More
UP000078540 UP000078542 UP000078541 UP000075809 UP000279307 UP000007755 UP000235965 UP000053097 UP000078492 UP000002358 UP000242457 UP000005203 UP000215335 UP000009046 UP000076502 UP000092461 UP000036403 UP000001819 UP000001292 UP000000803 UP000008711 UP000008792 UP000000304 UP000002282 UP000192221 UP000007801 UP000268350 UP000002320 UP000069940 UP000249989 UP000183832 UP000008820 UP000037069 UP000001070 UP000095300
UP000078540 UP000078542 UP000078541 UP000075809 UP000279307 UP000007755 UP000235965 UP000053097 UP000078492 UP000002358 UP000242457 UP000005203 UP000215335 UP000009046 UP000076502 UP000092461 UP000036403 UP000001819 UP000001292 UP000000803 UP000008711 UP000008792 UP000000304 UP000002282 UP000192221 UP000007801 UP000268350 UP000002320 UP000069940 UP000249989 UP000183832 UP000008820 UP000037069 UP000001070 UP000095300
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JG32
A0A2H1W8Z7
A0A212FJT3
A0A194PZP2
A0A194R9G0
A0A2A4JHI2
+ More
H9JKF1 E9I8Y7 A0A158NJR5 A0A195BFN7 A0A195C8F3 A0A195F243 A0A151WJA7 A0A3L8DVB6 F4X3X9 A0A2J7RTI3 A0A2J7RTI7 A0A026WPT9 A0A195EKP6 A0A0C9RRR6 K7IZK0 A0A310SPY5 A0A2A3EPY6 A0A088A7P6 A0A232FMY5 E0VX09 A0A154P9C2 A0A1L8DL91 A0A1L8DL04 A0A1B0CDI4 A0A1B6J9H9 A0A0J7L738 A0A1B6E836 A0A0C9R3I9 A0A1B6EBR3 A0A224XGN9 A0A0P4VHB2 A0A1L8DL93 Q297U8 B4HEN4 Q9VFK2 B3P0Q5 B4LY65 B4QZV4 Q8SY28 B4PST6 A0A1W4US81 B3LVG1 A0A3B0JIW8 B0W210 A0A182GUA4 A8WHF8 A0A0R3NHI5 A0A0Q9WNS7 A8JR05 A0A0R1E424 A0A1W4V4I9 A0A0N8P181 A0A0P8XWW0 A0A1J1HL51 A0A1Q3G475 A0A1Q3G469 Q17FX7 W8AV38 W8AR15 W8AJ15 W8BFR5 A0A0L0BKW0 A0A0K8V161 A0A0K8V6V7 A0A0A1XKZ3 A0A0K8UWK8 A0A0K8VI39 A0A0A1XEQ4 B4JTT0 A0A1I8QD82 W8AR19 A0A034V721
H9JKF1 E9I8Y7 A0A158NJR5 A0A195BFN7 A0A195C8F3 A0A195F243 A0A151WJA7 A0A3L8DVB6 F4X3X9 A0A2J7RTI3 A0A2J7RTI7 A0A026WPT9 A0A195EKP6 A0A0C9RRR6 K7IZK0 A0A310SPY5 A0A2A3EPY6 A0A088A7P6 A0A232FMY5 E0VX09 A0A154P9C2 A0A1L8DL91 A0A1L8DL04 A0A1B0CDI4 A0A1B6J9H9 A0A0J7L738 A0A1B6E836 A0A0C9R3I9 A0A1B6EBR3 A0A224XGN9 A0A0P4VHB2 A0A1L8DL93 Q297U8 B4HEN4 Q9VFK2 B3P0Q5 B4LY65 B4QZV4 Q8SY28 B4PST6 A0A1W4US81 B3LVG1 A0A3B0JIW8 B0W210 A0A182GUA4 A8WHF8 A0A0R3NHI5 A0A0Q9WNS7 A8JR05 A0A0R1E424 A0A1W4V4I9 A0A0N8P181 A0A0P8XWW0 A0A1J1HL51 A0A1Q3G475 A0A1Q3G469 Q17FX7 W8AV38 W8AR15 W8AJ15 W8BFR5 A0A0L0BKW0 A0A0K8V161 A0A0K8V6V7 A0A0A1XKZ3 A0A0K8UWK8 A0A0K8VI39 A0A0A1XEQ4 B4JTT0 A0A1I8QD82 W8AR19 A0A034V721
PDB
2PSO
E-value=1.1967e-48,
Score=491
Ontologies
GO
GO:0008289
GO:0007165
GO:0016021
GO:0007494
GO:0035024
GO:0007391
GO:0000902
GO:0035277
GO:0048814
GO:0008258
GO:0000912
GO:0030866
GO:0005938
GO:0051653
GO:0008586
GO:0007091
GO:0007274
GO:0007052
GO:0005096
GO:0000132
GO:0035160
GO:0007266
GO:0016323
GO:0007443
GO:0007424
GO:0030431
GO:0050806
GO:0035202
GO:0006468
GO:0005007
GO:0005524
GO:0008284
GO:0008543
GO:0004672
GO:0048384
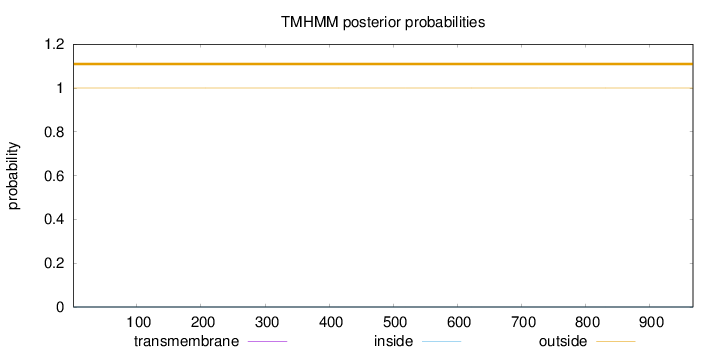
Topology
Length:
968
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00215
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00002
outside
1 - 968
Population Genetic Test Statistics
Pi
224.739045
Theta
183.887266
Tajima's D
0.831215
CLR
0.450834
CSRT
0.612919354032298
Interpretation
Uncertain
