Gene
KWMTBOMO03834
Pre Gene Modal
BGIBMGA010090
Annotation
PREDICTED:_fibroblast_growth_factor_receptor_isoform_X1_[Bombyx_mori]
Full name
Fibroblast growth factor receptor
Location in the cell
Cytoplasmic Reliability : 2.258 Nuclear Reliability : 1.744
Sequence
CDS
ATGGTGGAGTGGTTTAAAGATGGCCAGGCTCTGGATGGAGTCTCCAAGAGGATAACACAACAAAGGCAATGGTTAAGAATAAAAGGATTTAGAGCGAAAGACGCTGGTGTTTACTCGTGCCAAAACGAAGAAGATAAGTTCAAGGAAATGGCTGTCACGATCAAGCACAAGATTGGACAAGATCAAGATAATTTAGAAGAATATCAAGCAGATGTGGATATAGACAGGCCAATGCCAGAAATACTATCGCAACACCAAGCTGTTTCGCTACACGAAAATAACAAATTTGAAGAAAATACGACTGAAATAAAACGATTAGCAAGAACAATGAAATTAAAAGAAATCGATCCAAAAGATGAGGATCCTGATGATAAAAGTGAAAAGGATCATCCCGTTTACGGACACGTGGATGAAACTAAAAAGAAATACCCTCCCAAATTCAAACATCCATCAAAAATGTACACTATGGAAATGAAGCCTGCTGGCAGCTCTGTCAGATTTAAATGTCCTGCTGAAGGGAATCCCACGCCAAATATTACGTGGTACAAAAACAAAAGTAATCCAATACAAAGGACATACTTCCAGCCTAGCTACGCTAAATGGGCGATAAATTTAGATGAACTTACCAAAGCAGATAACGGAAATTACACTTGCGTTGTTTGCAATGAACTGGGTTGCATTGAGCACACTGTTGTGTTACACATACAAGAGCGCCTATACGCGAAGCCAGTTCTCACGCAAGGCGCAGTCAATCAAACTCGGATCGTAGGAGAGACGGCAAGATTCTCTTGCGATTTCCTCTCGGACATGCATCCGTATCTCTACTGGATGTATTTCTCGAAGGATGATTACGTCTATAGCGATGCCATCGATAGCGACGGCTCTGTTCTAGACTTAAATGACACGAGCAAATTCGTAACGAGCGATGATCCGTTTGACAAGTTGGAGCAGTTGACTTTGTACAACGTGACAAAAGAAGACGAAGGCTGGTACGTCTGCGTCGCTCTAAACACACTCGGCAATACAACCGCGAAAGGATACCTTACTGTTCTTGAATCTCCTCCGGAATTAGAGATACCGGATCACGGCAAGCACACGCTTCTAATCAACATACTGACCGCGGTACTGGGGGCTATGTTCTTCGTGGCCGCCATTATAGTGGTGATGGTCTTCAAGAAGCTAAAGCGAGAGAAGTTGAAGAAACAACTGGCAATAGAGACCGCGAGAGCTGTTATCGTTACGCACTGGACGAAGAAGGTTACAGTCGAGAAGCCACAGATGAACGGGTCTCCTAATACTACTGGAGAAGCTCTGCTGATGCCAGTCGTCAAAATCGAGAAGCAGAAACTGTCACAAGTACAGAATAATACATCGGACTCGATGATGATGTCGGAATACGAACTCCCAATGGACATAGACTGGGAAGTTCCCAGGGAATCGTTGTCGTTGGGAAAAGTCCTCGGCGAAGGGGAGTTCGGGAAGGTGGTGAAGGCTGAGTGCATCGGCATCCTGAAGCCGGGCTTGCAGTCCGTTGTTGCGGTCAAGATGTTGAAAGAGGGTCACACGGATGCTGAAATGATGGCTTTGGTGTCGGAAATGGAGATGATGAAGATGATAGGGAAACACGTAAACATAATCAACTTGTTGGGATGCTGCACGCAGGACGGACCTCTTTATGTCATAGTCGAGTACGCACCAAATGGAAACCTGAGGGAATTCCTGAGGAATCACAGACCCGGGAATAGGTACGAGTCACCAAACGAAGATTTGAAAGAAAAGAAGACGCTGACACAAAAAGATCTTGTATCTTTCTCCTATCAAGTGGCCAGAGGAATGGAGTATTTGGCTTCGAGACGATGCATCCATCGCGACTTGGCTGCTCGTAACGTGCTCGTGTCGGACGACTGCGTCCTCAAGATCGCCGACTTTGGTCTCGCTAAAGACGTCCACAGCAACGACTACTACAGGAAGAAGACAGAAGGGAGACTTCCCGTCCGGTGGATGGCGCCGGAGTCCTTGTACCACAAAGTGTTCACAACGCAGACCGACGTTTGGTCGTTTGGCGTGTTGCTATGGGAGATAATGACGCTGGGAGGCACCCCCTACCCCACCGTCCCCGGCCAGTACATGTACCAGCACCTCAGCGCCGGACACCGCATGGAGAAGCCGCCGTGCTGCAGTCTCGAGATTTACATGTTAATGCGCGAATGCTGGTCGTTTTCACCCGGCGACCGGCCGTCCTTCACCGAGCTGGTGGAGGATCTGGACAAGATTCTTACGGTCACAGCCAACCAGGAGTACCTGGACCTAGGCCTGCCACAGTTAGACACACCTCCTTCAAGTTACGACGGCTCCGGTGACGAAAGCGACGCTGAATTTCCATTTATAAAATAA
Protein
MVEWFKDGQALDGVSKRITQQRQWLRIKGFRAKDAGVYSCQNEEDKFKEMAVTIKHKIGQDQDNLEEYQADVDIDRPMPEILSQHQAVSLHENNKFEENTTEIKRLARTMKLKEIDPKDEDPDDKSEKDHPVYGHVDETKKKYPPKFKHPSKMYTMEMKPAGSSVRFKCPAEGNPTPNITWYKNKSNPIQRTYFQPSYAKWAINLDELTKADNGNYTCVVCNELGCIEHTVVLHIQERLYAKPVLTQGAVNQTRIVGETARFSCDFLSDMHPYLYWMYFSKDDYVYSDAIDSDGSVLDLNDTSKFVTSDDPFDKLEQLTLYNVTKEDEGWYVCVALNTLGNTTAKGYLTVLESPPELEIPDHGKHTLLINILTAVLGAMFFVAAIIVVMVFKKLKREKLKKQLAIETARAVIVTHWTKKVTVEKPQMNGSPNTTGEALLMPVVKIEKQKLSQVQNNTSDSMMMSEYELPMDIDWEVPRESLSLGKVLGEGEFGKVVKAECIGILKPGLQSVVAVKMLKEGHTDAEMMALVSEMEMMKMIGKHVNIINLLGCCTQDGPLYVIVEYAPNGNLREFLRNHRPGNRYESPNEDLKEKKTLTQKDLVSFSYQVARGMEYLASRRCIHRDLAARNVLVSDDCVLKIADFGLAKDVHSNDYYRKKTEGRLPVRWMAPESLYHKVFTTQTDVWSFGVLLWEIMTLGGTPYPTVPGQYMYQHLSAGHRMEKPPCCSLEIYMLMRECWSFSPGDRPSFTELVEDLDKILTVTANQEYLDLGLPQLDTPPSSYDGSGDESDAEFPFIK
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family. Fibroblast growth factor receptor subfamily.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Feature
chain Fibroblast growth factor receptor
Uniprot
Q1JUB8
A0A2A4J3S4
Q1JUB7
A0A1U9WTL1
A0A2A4J273
A0A2H1WDQ4
+ More
H9JKN9 A0A194QZK6 A0A194QDK7 A0A3S2LYN7 A0A212ESE0 A0A3L8E314 A0A0L7QMK7 A0A1W4XDH0 D6W6I2 A0A158NVU7 A0A0J7NYR0 A0A1W4X2U0 A0A195C7D7 A0A195BB67 A0A1B6K6L4 A0A1Y1KI53 F4WMU2 A0A0M9A011 A0A1Y1KD17 A0A026WLD5 A0A1B6C942 A0A224XJ43 A0A023F2X0 A0A0C9PZY2 A0A0C9QAF7 A0A2H8TF21 J9JQ68 A0A146M4S1 A0A1B6CPE6 A0A2S2PR88 A0A2S2Q3V2 A0A0U3IZJ2 E0VYK9 T1HVZ3 A0A154PKK4 A0A1J1J6U3 A0A182QYK6 A0A1J1J525 B0WJF5 A0A151J504 A0A1I8JVZ3 A0A182LC51 A0A088A4F4 A0A1S4GGZ1 Q5TTJ6 A0A182U4Y9 A0A2C9GQG7 A0A182UVU8 E9INR0 A0A195EWL7 A0A232EMX8 A0A151WPX0 A0A182LZD6 W5JVS5 A0A1I8JW45 A0A3F2YSE5 A0A1S4GGY3 F5HKZ1 Q17J18 E2A9R7 A0A182H093 A0A084VKS5 A0A067RG01 E9FS22 A0A0P6DKL4 A0A0P6ENA3 A0A0P5HSF2 A0A0P5VR39 A0A0P6AQB5 Q17BM1 A0A0P5DHL5 A0A0P5WVM3 A0A164YGT7 A0A0P6DJA9 A0A0P5U328 A0A1L8DJF7 A0A1L8DJG4 A0A0L0CAR6 A0A0K8VZG5 A0A034VJE0 A0A0A1WV42 A0A0P5CL91 A0A1I8M3C9 A0A1I8NXE3 A0A0P5TI12 A0A1I8M2A8 A0A0N8CJ71 A0A1L8DJR7 A0A131Z5C6 B4LZ96 A0A224YMV3 A0A147BNF1
H9JKN9 A0A194QZK6 A0A194QDK7 A0A3S2LYN7 A0A212ESE0 A0A3L8E314 A0A0L7QMK7 A0A1W4XDH0 D6W6I2 A0A158NVU7 A0A0J7NYR0 A0A1W4X2U0 A0A195C7D7 A0A195BB67 A0A1B6K6L4 A0A1Y1KI53 F4WMU2 A0A0M9A011 A0A1Y1KD17 A0A026WLD5 A0A1B6C942 A0A224XJ43 A0A023F2X0 A0A0C9PZY2 A0A0C9QAF7 A0A2H8TF21 J9JQ68 A0A146M4S1 A0A1B6CPE6 A0A2S2PR88 A0A2S2Q3V2 A0A0U3IZJ2 E0VYK9 T1HVZ3 A0A154PKK4 A0A1J1J6U3 A0A182QYK6 A0A1J1J525 B0WJF5 A0A151J504 A0A1I8JVZ3 A0A182LC51 A0A088A4F4 A0A1S4GGZ1 Q5TTJ6 A0A182U4Y9 A0A2C9GQG7 A0A182UVU8 E9INR0 A0A195EWL7 A0A232EMX8 A0A151WPX0 A0A182LZD6 W5JVS5 A0A1I8JW45 A0A3F2YSE5 A0A1S4GGY3 F5HKZ1 Q17J18 E2A9R7 A0A182H093 A0A084VKS5 A0A067RG01 E9FS22 A0A0P6DKL4 A0A0P6ENA3 A0A0P5HSF2 A0A0P5VR39 A0A0P6AQB5 Q17BM1 A0A0P5DHL5 A0A0P5WVM3 A0A164YGT7 A0A0P6DJA9 A0A0P5U328 A0A1L8DJF7 A0A1L8DJG4 A0A0L0CAR6 A0A0K8VZG5 A0A034VJE0 A0A0A1WV42 A0A0P5CL91 A0A1I8M3C9 A0A1I8NXE3 A0A0P5TI12 A0A1I8M2A8 A0A0N8CJ71 A0A1L8DJR7 A0A131Z5C6 B4LZ96 A0A224YMV3 A0A147BNF1
EC Number
2.7.10.1
Pubmed
16699027
19121390
26354079
22118469
30249741
18362917
+ More
19820115 21347285 28004739 21719571 24508170 25474469 26823975 20566863 20966253 12364791 21282665 28648823 20920257 23761445 17510324 20798317 26483478 24438588 24845553 21292972 26108605 25348373 25830018 25315136 26830274 17994087 18057021 28797301 29652888
19820115 21347285 28004739 21719571 24508170 25474469 26823975 20566863 20966253 12364791 21282665 28648823 20920257 23761445 17510324 20798317 26483478 24438588 24845553 21292972 26108605 25348373 25830018 25315136 26830274 17994087 18057021 28797301 29652888
EMBL
AB247566
BAE94421.1
NWSH01003526
PCG66164.1
AB247567
BAE94422.1
+ More
KX979914 AQY56784.1 PCG66165.1 ODYU01007976 SOQ51198.1 BABH01024924 KQ460930 KPJ10719.1 KQ459167 KPJ03504.1 RSAL01000112 RVE47023.1 AGBW02012819 OWR44397.1 QOIP01000001 RLU27094.1 KQ414885 KOC59858.1 KQ971307 EFA11574.2 ADTU01027496 LBMM01000766 KMQ97540.1 KQ978231 KYM96081.1 KQ976529 KYM81796.1 GECU01035110 GECU01000609 JAS72596.1 JAT07098.1 GEZM01086524 GEZM01086520 JAV59275.1 GL888221 EGI64476.1 KQ435803 KOX73103.1 GEZM01086523 GEZM01086521 JAV59274.1 KK107159 EZA56758.1 GEDC01027449 JAS09849.1 GFTR01008377 JAW08049.1 GBBI01003323 JAC15389.1 GBYB01007123 JAG76890.1 GBYB01011428 JAG81195.1 GFXV01000874 MBW12679.1 ABLF02028955 GDHC01004210 JAQ14419.1 GEDC01021946 JAS15352.1 GGMR01019235 MBY31854.1 GGMS01003088 MBY72291.1 KT355591 ALV82510.1 DS235845 EEB18465.1 ACPB03001692 ACPB03001693 ACPB03001694 ACPB03001695 ACPB03001696 KQ434948 KZC12382.1 CVRI01000072 CRL07508.1 AXCN02000226 CRL07509.1 DS231959 EDS29129.1 KQ980079 KYN17818.1 AAAB01008879 EAL40705.4 APCN01002106 GL764397 EFZ17715.1 KQ981940 KYN32670.1 NNAY01003260 OXU19719.1 KQ982850 KYQ49949.1 AXCM01000265 ADMH02000272 ETN67170.1 EGK96952.1 CH477235 EAT46693.1 GL437918 EFN69869.1 JXUM01100898 JXUM01100899 KQ564607 KXJ72033.1 ATLV01014246 KE524949 KFB38569.1 KK852498 KDR22672.1 GL732523 EFX89956.1 GDIQ01076139 JAN18598.1 GDIQ01061499 JAN33238.1 GDIQ01223583 JAK28142.1 GDIP01096690 JAM07025.1 GDIP01026044 JAM77671.1 CH477320 EAT43645.1 GDIP01172631 JAJ50771.1 GDIP01080986 JAM22729.1 LRGB01000915 KZS15239.1 GDIQ01076138 JAN18599.1 GDIP01120683 JAL83031.1 GFDF01007475 JAV06609.1 GFDF01007476 JAV06608.1 JRES01000760 KNC28549.1 GDHF01033965 GDHF01033128 GDHF01024275 GDHF01017768 GDHF01008011 GDHF01000710 GDHF01000440 JAI18349.1 JAI19186.1 JAI28039.1 JAI34546.1 JAI44303.1 JAI51604.1 JAI51874.1 GAKP01016339 GAKP01016338 JAC42614.1 GBXI01011917 JAD02375.1 GDIP01172630 JAJ50772.1 GDIP01126478 JAL77236.1 GDIP01126479 JAL77235.1 GFDF01007469 JAV06615.1 GEDV01003236 JAP85321.1 CH940650 EDW67103.2 KRF83103.1 GFPF01007801 MAA18947.1 GEGO01003432 JAR91972.1
KX979914 AQY56784.1 PCG66165.1 ODYU01007976 SOQ51198.1 BABH01024924 KQ460930 KPJ10719.1 KQ459167 KPJ03504.1 RSAL01000112 RVE47023.1 AGBW02012819 OWR44397.1 QOIP01000001 RLU27094.1 KQ414885 KOC59858.1 KQ971307 EFA11574.2 ADTU01027496 LBMM01000766 KMQ97540.1 KQ978231 KYM96081.1 KQ976529 KYM81796.1 GECU01035110 GECU01000609 JAS72596.1 JAT07098.1 GEZM01086524 GEZM01086520 JAV59275.1 GL888221 EGI64476.1 KQ435803 KOX73103.1 GEZM01086523 GEZM01086521 JAV59274.1 KK107159 EZA56758.1 GEDC01027449 JAS09849.1 GFTR01008377 JAW08049.1 GBBI01003323 JAC15389.1 GBYB01007123 JAG76890.1 GBYB01011428 JAG81195.1 GFXV01000874 MBW12679.1 ABLF02028955 GDHC01004210 JAQ14419.1 GEDC01021946 JAS15352.1 GGMR01019235 MBY31854.1 GGMS01003088 MBY72291.1 KT355591 ALV82510.1 DS235845 EEB18465.1 ACPB03001692 ACPB03001693 ACPB03001694 ACPB03001695 ACPB03001696 KQ434948 KZC12382.1 CVRI01000072 CRL07508.1 AXCN02000226 CRL07509.1 DS231959 EDS29129.1 KQ980079 KYN17818.1 AAAB01008879 EAL40705.4 APCN01002106 GL764397 EFZ17715.1 KQ981940 KYN32670.1 NNAY01003260 OXU19719.1 KQ982850 KYQ49949.1 AXCM01000265 ADMH02000272 ETN67170.1 EGK96952.1 CH477235 EAT46693.1 GL437918 EFN69869.1 JXUM01100898 JXUM01100899 KQ564607 KXJ72033.1 ATLV01014246 KE524949 KFB38569.1 KK852498 KDR22672.1 GL732523 EFX89956.1 GDIQ01076139 JAN18598.1 GDIQ01061499 JAN33238.1 GDIQ01223583 JAK28142.1 GDIP01096690 JAM07025.1 GDIP01026044 JAM77671.1 CH477320 EAT43645.1 GDIP01172631 JAJ50771.1 GDIP01080986 JAM22729.1 LRGB01000915 KZS15239.1 GDIQ01076138 JAN18599.1 GDIP01120683 JAL83031.1 GFDF01007475 JAV06609.1 GFDF01007476 JAV06608.1 JRES01000760 KNC28549.1 GDHF01033965 GDHF01033128 GDHF01024275 GDHF01017768 GDHF01008011 GDHF01000710 GDHF01000440 JAI18349.1 JAI19186.1 JAI28039.1 JAI34546.1 JAI44303.1 JAI51604.1 JAI51874.1 GAKP01016339 GAKP01016338 JAC42614.1 GBXI01011917 JAD02375.1 GDIP01172630 JAJ50772.1 GDIP01126478 JAL77236.1 GDIP01126479 JAL77235.1 GFDF01007469 JAV06615.1 GEDV01003236 JAP85321.1 CH940650 EDW67103.2 KRF83103.1 GFPF01007801 MAA18947.1 GEGO01003432 JAR91972.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000279307 UP000053825 UP000192223 UP000007266 UP000005205 UP000036403 UP000078542 UP000078540 UP000007755 UP000053105 UP000053097 UP000007819 UP000009046 UP000015103 UP000076502 UP000183832 UP000075886 UP000002320 UP000078492 UP000076407 UP000075882 UP000005203 UP000007062 UP000075902 UP000075840 UP000075903 UP000078541 UP000215335 UP000075809 UP000075883 UP000000673 UP000008820 UP000000311 UP000069940 UP000249989 UP000030765 UP000027135 UP000000305 UP000076858 UP000037069 UP000095301 UP000095300 UP000008792
UP000279307 UP000053825 UP000192223 UP000007266 UP000005205 UP000036403 UP000078542 UP000078540 UP000007755 UP000053105 UP000053097 UP000007819 UP000009046 UP000015103 UP000076502 UP000183832 UP000075886 UP000002320 UP000078492 UP000076407 UP000075882 UP000005203 UP000007062 UP000075902 UP000075840 UP000075903 UP000078541 UP000215335 UP000075809 UP000075883 UP000000673 UP000008820 UP000000311 UP000069940 UP000249989 UP000030765 UP000027135 UP000000305 UP000076858 UP000037069 UP000095301 UP000095300 UP000008792
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR013098 Ig_I-set
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR016248 FGF_rcpt_fam
IPR036179 Ig-like_dom_sf
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR013151 Immunoglobulin
IPR013083 Znf_RING/FYVE/PHD
IPR011016 Znf_RING-CH
IPR001005 SANT/Myb
IPR009057 Homeobox-like_sf
IPR017884 SANT_dom
IPR037020 Hemocyanin_C_sf
IPR003959 ATPase_AAA_core
IPR015342 PEX-N_psi_beta-barrel
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR005204 Hemocyanin_N
IPR014756 Ig_E-set
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR009010 Asp_de-COase-like_dom_sf
IPR036697 Hemocyanin_N_sf
IPR029067 CDC48_domain_2-like_sf
IPR005203 Hemocyanin_C
IPR013106 Ig_V-set
IPR013098 Ig_I-set
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR017441 Protein_kinase_ATP_BS
IPR013783 Ig-like_fold
IPR003598 Ig_sub2
IPR016248 FGF_rcpt_fam
IPR036179 Ig-like_dom_sf
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR013151 Immunoglobulin
IPR013083 Znf_RING/FYVE/PHD
IPR011016 Znf_RING-CH
IPR001005 SANT/Myb
IPR009057 Homeobox-like_sf
IPR017884 SANT_dom
IPR037020 Hemocyanin_C_sf
IPR003959 ATPase_AAA_core
IPR015342 PEX-N_psi_beta-barrel
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR005204 Hemocyanin_N
IPR014756 Ig_E-set
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR009010 Asp_de-COase-like_dom_sf
IPR036697 Hemocyanin_N_sf
IPR029067 CDC48_domain_2-like_sf
IPR005203 Hemocyanin_C
IPR013106 Ig_V-set
SUPFAM
Gene 3D
ProteinModelPortal
Q1JUB8
A0A2A4J3S4
Q1JUB7
A0A1U9WTL1
A0A2A4J273
A0A2H1WDQ4
+ More
H9JKN9 A0A194QZK6 A0A194QDK7 A0A3S2LYN7 A0A212ESE0 A0A3L8E314 A0A0L7QMK7 A0A1W4XDH0 D6W6I2 A0A158NVU7 A0A0J7NYR0 A0A1W4X2U0 A0A195C7D7 A0A195BB67 A0A1B6K6L4 A0A1Y1KI53 F4WMU2 A0A0M9A011 A0A1Y1KD17 A0A026WLD5 A0A1B6C942 A0A224XJ43 A0A023F2X0 A0A0C9PZY2 A0A0C9QAF7 A0A2H8TF21 J9JQ68 A0A146M4S1 A0A1B6CPE6 A0A2S2PR88 A0A2S2Q3V2 A0A0U3IZJ2 E0VYK9 T1HVZ3 A0A154PKK4 A0A1J1J6U3 A0A182QYK6 A0A1J1J525 B0WJF5 A0A151J504 A0A1I8JVZ3 A0A182LC51 A0A088A4F4 A0A1S4GGZ1 Q5TTJ6 A0A182U4Y9 A0A2C9GQG7 A0A182UVU8 E9INR0 A0A195EWL7 A0A232EMX8 A0A151WPX0 A0A182LZD6 W5JVS5 A0A1I8JW45 A0A3F2YSE5 A0A1S4GGY3 F5HKZ1 Q17J18 E2A9R7 A0A182H093 A0A084VKS5 A0A067RG01 E9FS22 A0A0P6DKL4 A0A0P6ENA3 A0A0P5HSF2 A0A0P5VR39 A0A0P6AQB5 Q17BM1 A0A0P5DHL5 A0A0P5WVM3 A0A164YGT7 A0A0P6DJA9 A0A0P5U328 A0A1L8DJF7 A0A1L8DJG4 A0A0L0CAR6 A0A0K8VZG5 A0A034VJE0 A0A0A1WV42 A0A0P5CL91 A0A1I8M3C9 A0A1I8NXE3 A0A0P5TI12 A0A1I8M2A8 A0A0N8CJ71 A0A1L8DJR7 A0A131Z5C6 B4LZ96 A0A224YMV3 A0A147BNF1
H9JKN9 A0A194QZK6 A0A194QDK7 A0A3S2LYN7 A0A212ESE0 A0A3L8E314 A0A0L7QMK7 A0A1W4XDH0 D6W6I2 A0A158NVU7 A0A0J7NYR0 A0A1W4X2U0 A0A195C7D7 A0A195BB67 A0A1B6K6L4 A0A1Y1KI53 F4WMU2 A0A0M9A011 A0A1Y1KD17 A0A026WLD5 A0A1B6C942 A0A224XJ43 A0A023F2X0 A0A0C9PZY2 A0A0C9QAF7 A0A2H8TF21 J9JQ68 A0A146M4S1 A0A1B6CPE6 A0A2S2PR88 A0A2S2Q3V2 A0A0U3IZJ2 E0VYK9 T1HVZ3 A0A154PKK4 A0A1J1J6U3 A0A182QYK6 A0A1J1J525 B0WJF5 A0A151J504 A0A1I8JVZ3 A0A182LC51 A0A088A4F4 A0A1S4GGZ1 Q5TTJ6 A0A182U4Y9 A0A2C9GQG7 A0A182UVU8 E9INR0 A0A195EWL7 A0A232EMX8 A0A151WPX0 A0A182LZD6 W5JVS5 A0A1I8JW45 A0A3F2YSE5 A0A1S4GGY3 F5HKZ1 Q17J18 E2A9R7 A0A182H093 A0A084VKS5 A0A067RG01 E9FS22 A0A0P6DKL4 A0A0P6ENA3 A0A0P5HSF2 A0A0P5VR39 A0A0P6AQB5 Q17BM1 A0A0P5DHL5 A0A0P5WVM3 A0A164YGT7 A0A0P6DJA9 A0A0P5U328 A0A1L8DJF7 A0A1L8DJG4 A0A0L0CAR6 A0A0K8VZG5 A0A034VJE0 A0A0A1WV42 A0A0P5CL91 A0A1I8M3C9 A0A1I8NXE3 A0A0P5TI12 A0A1I8M2A8 A0A0N8CJ71 A0A1L8DJR7 A0A131Z5C6 B4LZ96 A0A224YMV3 A0A147BNF1
PDB
3GQI
E-value=4.58006e-116,
Score=1072
Ontologies
GO
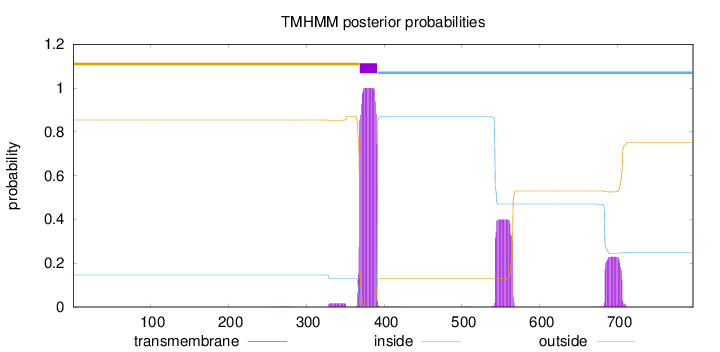
Topology
Subcellular location
Nucleus
Length:
797
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.9403399999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.14633
outside
1 - 368
TMhelix
369 - 391
inside
392 - 797
Population Genetic Test Statistics
Pi
244.355745
Theta
168.68045
Tajima's D
1.138418
CLR
0.561531
CSRT
0.697365131743413
Interpretation
Uncertain
