Pre Gene Modal
BGIBMGA010004
Annotation
syntaxin_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.537
Sequence
CDS
ATGTCAGAACGAGAACCAATTAATGGTTCAGGGCGAGACTATGGAGCCACAGCCTCTGTGCCGGCTGTGGGCTTTGCAGATTTCAGTCCAACGGAACTCTACAATTTGAGTGAGGGCATCGCAGAGAATGTCAATACGATTAACAGTGGACTACTGTCGCTTGAGAAGATGATGAAGCAAATCGGTGGCCCAAATGATAGCGTCCAACTTAGAGATAAAATGTAA
Protein
MSEREPINGSGRDYGATASVPAVGFADFSPTELYNLSEGIAENVNTINSGLLSLEKMMKQIGGPNDSVQLRDKM
Summary
Similarity
Belongs to the syntaxin family.
Uniprot
Q1HPH0
H9JKF3
A0A2A4JKA8
A0A2H1WV58
A0A194QD83
A0A194QZ32
+ More
A0A437B9F8 A0A1E1VXP0 A0A212EJ65 S4PD96 A0A1A9VWC7 A0A1B0GD27 A0A1A9ZA37 A0A067RLD3 A0A2J7PQ42 A0A1A9WGB3 A0A1A9Y3D6 A0A1B0BCH5 E2C0V8 A0A2P8Z5T7 T1PH92 B4J0S1 A0A232ERJ0 B4LC11 A0A1I8N0A2 A0A1I8P011 W8CAY1 A0A1L8EI63 A0A1B6GFX6 A0A026W268 A0A1W4VIY8 B4HGP1 B4QIZ1 A0A0M9A5J7 E2AKD8 A0A2A3EM18 U5EIB6 A0A0A1WS00 A0A0K8UVU1 A0A034WWI3 A0A0C9RSY3 B4GS63 Q2M037 A0A1L8DUJ9 B4PHB9 A0A0J7KJY0 A0A1L8DUJ4 A0A0K8TNL4 V9IHS1 A0A2A3EKJ8 A0A088A431 A0A154NW01 A0A1B0DKU0 Q9VU45 B3MAZ2 A0A151ID19 A0A158N9M4 B3NCZ1 A0A0L0CFB2 A0A151ISU6 A0A0L7R603 A0A1B6DV26 A0A195BNJ9 F4WTR4 A0A195F9E4 A0A1B6LMN7 B0X748 A0A3B0JSU3 A0A0P4VT18 A0A3B0J4G4 A0A151XGT0 A0A069DRT5 A0A224XUE1 A0A0V0GD30 A0A336MRT9 R4V148 A0A084VBY0 B4KZS1 A0A1Q3FD80 B4MLZ1 Q16VZ6 A0A182QP07 A0A182NPP4 A0A0P4W7A0 A0A182PEJ2 A0A182M4P8 A0A182R4F5 A0A182JVR5 A0A182WED4 A0A1Q3FDR9 W5J6K3 A0A023EMU8 A0A182G2N5 A0A182JG91 A0A023EPB0 A0A182YL49 A0A182SZ69 A0A1D2NIX5
A0A437B9F8 A0A1E1VXP0 A0A212EJ65 S4PD96 A0A1A9VWC7 A0A1B0GD27 A0A1A9ZA37 A0A067RLD3 A0A2J7PQ42 A0A1A9WGB3 A0A1A9Y3D6 A0A1B0BCH5 E2C0V8 A0A2P8Z5T7 T1PH92 B4J0S1 A0A232ERJ0 B4LC11 A0A1I8N0A2 A0A1I8P011 W8CAY1 A0A1L8EI63 A0A1B6GFX6 A0A026W268 A0A1W4VIY8 B4HGP1 B4QIZ1 A0A0M9A5J7 E2AKD8 A0A2A3EM18 U5EIB6 A0A0A1WS00 A0A0K8UVU1 A0A034WWI3 A0A0C9RSY3 B4GS63 Q2M037 A0A1L8DUJ9 B4PHB9 A0A0J7KJY0 A0A1L8DUJ4 A0A0K8TNL4 V9IHS1 A0A2A3EKJ8 A0A088A431 A0A154NW01 A0A1B0DKU0 Q9VU45 B3MAZ2 A0A151ID19 A0A158N9M4 B3NCZ1 A0A0L0CFB2 A0A151ISU6 A0A0L7R603 A0A1B6DV26 A0A195BNJ9 F4WTR4 A0A195F9E4 A0A1B6LMN7 B0X748 A0A3B0JSU3 A0A0P4VT18 A0A3B0J4G4 A0A151XGT0 A0A069DRT5 A0A224XUE1 A0A0V0GD30 A0A336MRT9 R4V148 A0A084VBY0 B4KZS1 A0A1Q3FD80 B4MLZ1 Q16VZ6 A0A182QP07 A0A182NPP4 A0A0P4W7A0 A0A182PEJ2 A0A182M4P8 A0A182R4F5 A0A182JVR5 A0A182WED4 A0A1Q3FDR9 W5J6K3 A0A023EMU8 A0A182G2N5 A0A182JG91 A0A023EPB0 A0A182YL49 A0A182SZ69 A0A1D2NIX5
Pubmed
19121390
26354079
22118469
23622113
24845553
20798317
+ More
29403074 17994087 28648823 18057021 25315136 24495485 24508170 30249741 22936249 25830018 25348373 15632085 23185243 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 26108605 21719571 27129103 26334808 24438588 17510324 20920257 23761445 24945155 26483478 25244985 27289101
29403074 17994087 28648823 18057021 25315136 24495485 24508170 30249741 22936249 25830018 25348373 15632085 23185243 17550304 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 26108605 21719571 27129103 26334808 24438588 17510324 20920257 23761445 24945155 26483478 25244985 27289101
EMBL
DQ443432
ABF51521.1
BABH01024910
BABH01024911
NWSH01001172
PCG72259.1
+ More
ODYU01011304 SOQ56955.1 KQ459167 KPJ03503.1 KQ460930 KPJ10717.1 RSAL01000112 RVE47022.1 GDQN01011610 GDQN01001128 JAT79444.1 JAT89926.1 AGBW02014515 OWR41543.1 GAIX01007595 JAA84965.1 CCAG010002291 KK852442 KDR23848.1 NEVH01022640 PNF18454.1 JXJN01012002 GL451850 EFN78394.1 PYGN01000183 PSN51863.1 KA647263 AFP61892.1 CH916366 EDV97926.1 NNAY01002583 OXU20993.1 CH940647 EDW69811.1 KRF84596.1 GAMC01000954 JAC05602.1 GFDG01000454 JAV18345.1 GECZ01008440 JAS61329.1 KK107502 QOIP01000014 EZA49691.1 RLU14882.1 CH480815 EDW41349.1 CM000363 CM002912 EDX10322.1 KMY99358.1 KQ435737 KOX76906.1 GL440262 EFN66080.1 KZ288219 PBC32246.1 GANO01002692 JAB57179.1 GBXI01012478 JAD01814.1 GDHF01021661 GDHF01005359 JAI30653.1 JAI46955.1 GAKP01000794 JAC58158.1 GBYB01011740 JAG81507.1 CH479188 EDW40598.1 CH379069 EAL31099.1 KRT07868.1 KRT07869.1 GFDF01003981 JAV10103.1 CM000159 EDW94380.1 LBMM01006279 KMQ90743.1 GFDF01003980 JAV10104.1 GDAI01001875 JAI15728.1 JR046479 AEY60202.1 PBC32247.1 KQ434772 KZC03865.1 AJVK01016112 AJVK01016113 AE014296 AY069551 AAF49845.1 AAL39696.1 AGB94487.1 CH902618 EDV40258.1 KQ977991 KYM98304.1 ADTU01009698 CH954178 EDV51647.1 JRES01000484 KNC30930.1 KQ981064 KYN09906.1 KQ414648 KOC66249.1 GEDC01016353 GEDC01007809 JAS20945.1 JAS29489.1 KQ976428 KYM87797.1 GL888344 EGI62379.1 KQ981727 KYN36674.1 GEBQ01014985 JAT24992.1 DS232438 EDS41742.1 OUUW01000002 SPP76426.1 GDKW01000247 JAI56348.1 SPP76427.1 KQ982169 KYQ59511.1 GBGD01002503 JAC86386.1 GFTR01004703 JAW11723.1 GECL01000374 JAP05750.1 UFQT01002152 SSX32780.1 KC571868 AGM32367.1 ATLV01009822 KE524548 KFB35474.1 CH933809 EDW19027.1 GFDL01009520 JAV25525.1 CH963847 EDW73202.1 CH477579 EAT38717.1 AXCN02001655 GDRN01080729 GDRN01080728 JAI62150.1 AXCM01001368 GFDL01009407 JAV25638.1 ADMH02001980 ETN60092.1 GAPW01002911 JAC10687.1 JXUM01139743 KQ568947 KXJ68828.1 GAPW01002767 JAC10831.1 LJIJ01000028 ODN05201.1
ODYU01011304 SOQ56955.1 KQ459167 KPJ03503.1 KQ460930 KPJ10717.1 RSAL01000112 RVE47022.1 GDQN01011610 GDQN01001128 JAT79444.1 JAT89926.1 AGBW02014515 OWR41543.1 GAIX01007595 JAA84965.1 CCAG010002291 KK852442 KDR23848.1 NEVH01022640 PNF18454.1 JXJN01012002 GL451850 EFN78394.1 PYGN01000183 PSN51863.1 KA647263 AFP61892.1 CH916366 EDV97926.1 NNAY01002583 OXU20993.1 CH940647 EDW69811.1 KRF84596.1 GAMC01000954 JAC05602.1 GFDG01000454 JAV18345.1 GECZ01008440 JAS61329.1 KK107502 QOIP01000014 EZA49691.1 RLU14882.1 CH480815 EDW41349.1 CM000363 CM002912 EDX10322.1 KMY99358.1 KQ435737 KOX76906.1 GL440262 EFN66080.1 KZ288219 PBC32246.1 GANO01002692 JAB57179.1 GBXI01012478 JAD01814.1 GDHF01021661 GDHF01005359 JAI30653.1 JAI46955.1 GAKP01000794 JAC58158.1 GBYB01011740 JAG81507.1 CH479188 EDW40598.1 CH379069 EAL31099.1 KRT07868.1 KRT07869.1 GFDF01003981 JAV10103.1 CM000159 EDW94380.1 LBMM01006279 KMQ90743.1 GFDF01003980 JAV10104.1 GDAI01001875 JAI15728.1 JR046479 AEY60202.1 PBC32247.1 KQ434772 KZC03865.1 AJVK01016112 AJVK01016113 AE014296 AY069551 AAF49845.1 AAL39696.1 AGB94487.1 CH902618 EDV40258.1 KQ977991 KYM98304.1 ADTU01009698 CH954178 EDV51647.1 JRES01000484 KNC30930.1 KQ981064 KYN09906.1 KQ414648 KOC66249.1 GEDC01016353 GEDC01007809 JAS20945.1 JAS29489.1 KQ976428 KYM87797.1 GL888344 EGI62379.1 KQ981727 KYN36674.1 GEBQ01014985 JAT24992.1 DS232438 EDS41742.1 OUUW01000002 SPP76426.1 GDKW01000247 JAI56348.1 SPP76427.1 KQ982169 KYQ59511.1 GBGD01002503 JAC86386.1 GFTR01004703 JAW11723.1 GECL01000374 JAP05750.1 UFQT01002152 SSX32780.1 KC571868 AGM32367.1 ATLV01009822 KE524548 KFB35474.1 CH933809 EDW19027.1 GFDL01009520 JAV25525.1 CH963847 EDW73202.1 CH477579 EAT38717.1 AXCN02001655 GDRN01080729 GDRN01080728 JAI62150.1 AXCM01001368 GFDL01009407 JAV25638.1 ADMH02001980 ETN60092.1 GAPW01002911 JAC10687.1 JXUM01139743 KQ568947 KXJ68828.1 GAPW01002767 JAC10831.1 LJIJ01000028 ODN05201.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000078200 UP000092444 UP000092445 UP000027135 UP000235965 UP000091820 UP000092443 UP000092460 UP000008237 UP000245037 UP000001070 UP000215335 UP000008792 UP000095301 UP000095300 UP000053097 UP000279307 UP000192221 UP000001292 UP000000304 UP000053105 UP000000311 UP000242457 UP000008744 UP000001819 UP000002282 UP000036403 UP000005203 UP000076502 UP000092462 UP000000803 UP000007801 UP000078542 UP000005205 UP000008711 UP000037069 UP000078492 UP000053825 UP000078540 UP000007755 UP000078541 UP000002320 UP000268350 UP000075809 UP000030765 UP000009192 UP000007798 UP000008820 UP000075886 UP000075884 UP000075885 UP000075883 UP000075900 UP000075881 UP000075920 UP000000673 UP000069940 UP000249989 UP000075880 UP000076408 UP000075901 UP000094527
UP000078200 UP000092444 UP000092445 UP000027135 UP000235965 UP000091820 UP000092443 UP000092460 UP000008237 UP000245037 UP000001070 UP000215335 UP000008792 UP000095301 UP000095300 UP000053097 UP000279307 UP000192221 UP000001292 UP000000304 UP000053105 UP000000311 UP000242457 UP000008744 UP000001819 UP000002282 UP000036403 UP000005203 UP000076502 UP000092462 UP000000803 UP000007801 UP000078542 UP000005205 UP000008711 UP000037069 UP000078492 UP000053825 UP000078540 UP000007755 UP000078541 UP000002320 UP000268350 UP000075809 UP000030765 UP000009192 UP000007798 UP000008820 UP000075886 UP000075884 UP000075885 UP000075883 UP000075900 UP000075881 UP000075920 UP000000673 UP000069940 UP000249989 UP000075880 UP000076408 UP000075901 UP000094527
Interpro
SUPFAM
SSF47661
SSF47661
ProteinModelPortal
Q1HPH0
H9JKF3
A0A2A4JKA8
A0A2H1WV58
A0A194QD83
A0A194QZ32
+ More
A0A437B9F8 A0A1E1VXP0 A0A212EJ65 S4PD96 A0A1A9VWC7 A0A1B0GD27 A0A1A9ZA37 A0A067RLD3 A0A2J7PQ42 A0A1A9WGB3 A0A1A9Y3D6 A0A1B0BCH5 E2C0V8 A0A2P8Z5T7 T1PH92 B4J0S1 A0A232ERJ0 B4LC11 A0A1I8N0A2 A0A1I8P011 W8CAY1 A0A1L8EI63 A0A1B6GFX6 A0A026W268 A0A1W4VIY8 B4HGP1 B4QIZ1 A0A0M9A5J7 E2AKD8 A0A2A3EM18 U5EIB6 A0A0A1WS00 A0A0K8UVU1 A0A034WWI3 A0A0C9RSY3 B4GS63 Q2M037 A0A1L8DUJ9 B4PHB9 A0A0J7KJY0 A0A1L8DUJ4 A0A0K8TNL4 V9IHS1 A0A2A3EKJ8 A0A088A431 A0A154NW01 A0A1B0DKU0 Q9VU45 B3MAZ2 A0A151ID19 A0A158N9M4 B3NCZ1 A0A0L0CFB2 A0A151ISU6 A0A0L7R603 A0A1B6DV26 A0A195BNJ9 F4WTR4 A0A195F9E4 A0A1B6LMN7 B0X748 A0A3B0JSU3 A0A0P4VT18 A0A3B0J4G4 A0A151XGT0 A0A069DRT5 A0A224XUE1 A0A0V0GD30 A0A336MRT9 R4V148 A0A084VBY0 B4KZS1 A0A1Q3FD80 B4MLZ1 Q16VZ6 A0A182QP07 A0A182NPP4 A0A0P4W7A0 A0A182PEJ2 A0A182M4P8 A0A182R4F5 A0A182JVR5 A0A182WED4 A0A1Q3FDR9 W5J6K3 A0A023EMU8 A0A182G2N5 A0A182JG91 A0A023EPB0 A0A182YL49 A0A182SZ69 A0A1D2NIX5
A0A437B9F8 A0A1E1VXP0 A0A212EJ65 S4PD96 A0A1A9VWC7 A0A1B0GD27 A0A1A9ZA37 A0A067RLD3 A0A2J7PQ42 A0A1A9WGB3 A0A1A9Y3D6 A0A1B0BCH5 E2C0V8 A0A2P8Z5T7 T1PH92 B4J0S1 A0A232ERJ0 B4LC11 A0A1I8N0A2 A0A1I8P011 W8CAY1 A0A1L8EI63 A0A1B6GFX6 A0A026W268 A0A1W4VIY8 B4HGP1 B4QIZ1 A0A0M9A5J7 E2AKD8 A0A2A3EM18 U5EIB6 A0A0A1WS00 A0A0K8UVU1 A0A034WWI3 A0A0C9RSY3 B4GS63 Q2M037 A0A1L8DUJ9 B4PHB9 A0A0J7KJY0 A0A1L8DUJ4 A0A0K8TNL4 V9IHS1 A0A2A3EKJ8 A0A088A431 A0A154NW01 A0A1B0DKU0 Q9VU45 B3MAZ2 A0A151ID19 A0A158N9M4 B3NCZ1 A0A0L0CFB2 A0A151ISU6 A0A0L7R603 A0A1B6DV26 A0A195BNJ9 F4WTR4 A0A195F9E4 A0A1B6LMN7 B0X748 A0A3B0JSU3 A0A0P4VT18 A0A3B0J4G4 A0A151XGT0 A0A069DRT5 A0A224XUE1 A0A0V0GD30 A0A336MRT9 R4V148 A0A084VBY0 B4KZS1 A0A1Q3FD80 B4MLZ1 Q16VZ6 A0A182QP07 A0A182NPP4 A0A0P4W7A0 A0A182PEJ2 A0A182M4P8 A0A182R4F5 A0A182JVR5 A0A182WED4 A0A1Q3FDR9 W5J6K3 A0A023EMU8 A0A182G2N5 A0A182JG91 A0A023EPB0 A0A182YL49 A0A182SZ69 A0A1D2NIX5
Ontologies
GO
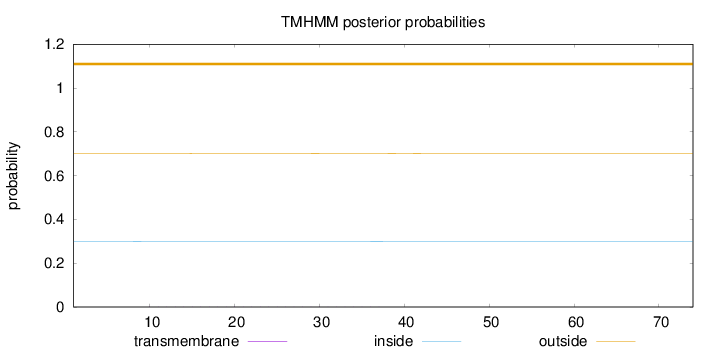
Topology
Length:
74
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02247
Exp number, first 60 AAs:
0.02247
Total prob of N-in:
0.29867
outside
1 - 74
Population Genetic Test Statistics
Pi
310.277292
Theta
188.95792
Tajima's D
1.674793
CLR
0
CSRT
0.829158542072896
Interpretation
Uncertain
