Gene
KWMTBOMO03831
Pre Gene Modal
BGIBMGA010089
Annotation
PREDICTED:_general_transcription_factor_IIH_subunit_1_[Bombyx_mori]
Full name
General transcription factor IIH subunit 1
Alternative Name
TFIIH basal transcription factor complex subunit 1
Location in the cell
Nuclear Reliability : 3.831
Sequence
CDS
ATGACAACATCATCCGAAGATGTTTTGCTCAGCGTGAATTGCGTACGCTATAAGAAGGGAGACGGGACACTGTATGTAATGAATCAAAGATTAGCATGGATGTTGGAACACAGAGACACAGTCGCAGTATCTCATAAATATGCTGATATCAAAACACAAAAAATTTCACCAGCTGGCAAGGCTAAAATACAGCTTCAAGTGGTTTTACATGATGGCACATGTTCTACATTTCACTTTGTTAACCCGGGTGGCCCAGAGGCTCAGGCCAAAGACAGAGATCAAGTGATGGTATTACTACAAAACCTTTTACCAAAGTTCAAAAGACAGATTGATGGTGAACTAGAAATGAAATCTAAATTGTTGTCATTGCATCCAACATTAAAACATTTGTACGAGGATTTAGTTATATCTAAAGTTATTAATAGTGAGGAATACTGGAATACTCCTACTTTGAAACAGTATACAGAATCTAGTAACATTAAACAGGAAGCTGGTGTGTCAGGTGCATTCCTTGCTGATATTCAACCACAGACAGACGGTTGCAATGGTTTGAAATACAATCTAACTCAAGATATCATAGATGCTATATTCAAAACATATCCAGCAGTCAGAAAGAAACATATTGATTATGTACCCAATAAAATGACGGAAGCTGAATTCTGGACGAAATTCTTTCAATCACATTATTTTCACAGAGATCGAATAGCTTCGTCATCGAGCAAGGATTTATTTGGAGAATGTGCTAAATTAGATGACCACGCTATTGCTTCAGCAATGAGACACACAACACTAGATCTTACGGTTGATTTGAATAAGTTCATAGAGCACGTTCCACTGTTACCGACTGAAGAAGAAGCTAGCAATGAGAAGGATCGATCTGATAATAATTCTATACACAGAAATATGATTAAACGTTTCAACCAACACTCTATTATGGTGCTCAAAGCTAGTCAAAAATCAAATTCTAGTAATAATAGTAATAGCAGTAGTAGTAAAACATCGAATAACAACAATAAAGTTATCGAAAACGGTGTCAAAGATACAAATGGTGTCTCAAAGAGACCGGCCGAACACAGTACAGAACCGATAGAAAAGAAAAAACGAATATTAGAGCAGATACACTACGAGGATCTAGTTGCAGACCAGGGAAATGATGATGTACAGGAATTAAAATTGTCTAAGGTAGAAAGATATTTATTAGGACCGTCGTATCAGATAAGTCAGACGAGTGTTCAGACGAGCCACCCGCCGCCGCTGTCGGCCTTGGCGTCCGTATGTCAGGCGTGGTCGAGCGGCCAACAGTGCCAGCGGCCGGCGCGGGTGAGCGCGGGCGCGGCTGTGGGCGCGCTGGGGGAGCTCAGTCCGGGCGGCGCGCTCATGCGGCACCACCACGCCGCCTCTATGGCACAACTAATACCGGCGGACGTGAAGGCGGAGCTTCAGAGGCTGTACGCGTCGTGCGGCGAGCTGGCGAGGGAGCTTTGGCGCAGCTTCCCGCAGCCGGGCGCGCCGCCAGCCAACGAGCAGGACGCGGCGCCGCGGGCGACCATGTTCTACGACGCGCTGTTGCGGTTTAGGAATTTGAAACTGAGGCCCTTCGAGGAGAAGATGCTCCGTGACCTGACGCCCCTGGCGACGACTCTAACGAAACATTTGAACCAAATAATAGACACGGCCTGCGCTAAGTACGTGCTCTGGCAACAGAGGCAAGCGAAGATACGGTAG
Protein
MTTSSEDVLLSVNCVRYKKGDGTLYVMNQRLAWMLEHRDTVAVSHKYADIKTQKISPAGKAKIQLQVVLHDGTCSTFHFVNPGGPEAQAKDRDQVMVLLQNLLPKFKRQIDGELEMKSKLLSLHPTLKHLYEDLVISKVINSEEYWNTPTLKQYTESSNIKQEAGVSGAFLADIQPQTDGCNGLKYNLTQDIIDAIFKTYPAVRKKHIDYVPNKMTEAEFWTKFFQSHYFHRDRIASSSSKDLFGECAKLDDHAIASAMRHTTLDLTVDLNKFIEHVPLLPTEEEASNEKDRSDNNSIHRNMIKRFNQHSIMVLKASQKSNSSNNSNSSSSKTSNNNNKVIENGVKDTNGVSKRPAEHSTEPIEKKKRILEQIHYEDLVADQGNDDVQELKLSKVERYLLGPSYQISQTSVQTSHPPPLSALASVCQAWSSGQQCQRPARVSAGAAVGALGELSPGGALMRHHHAASMAQLIPADVKAELQRLYASCGELARELWRSFPQPGAPPANEQDAAPRATMFYDALLRFRNLKLRPFEEKMLRDLTPLATTLTKHLNQIIDTACAKYVLWQQRQAKIR
Summary
Description
Component of the general transcription and DNA repair factor IIH (TFIIH) core complex, which is involved in general and transcription-coupled nucleotide excision repair (NER) of damaged DNA and, when complexed to CAK, in RNA transcription by RNA polymerase II. In NER, TFIIH acts by opening DNA around the lesion to allow the excision of the damaged oligonucleotide and its replacement by a new DNA fragment. In transcription, TFIIH has an essential role in transcription initiation. When the pre-initiation complex (PIC) has been established, TFIIH is required for promoter opening and promoter escape. Phosphorylation of the C-terminal tail (CTD) of the largest subunit of RNA polymerase II by the kinase module CAK controls the initiation of transcription.
Subunit
Component of the 7-subunit TFIIH core complex composed of XPB/ERCC3, XPD/ERCC2, GTF2H1, GTF2H2, GTF2H3, GTF2H4 and GTF2H5, which is active in NER. The core complex associates with the 3-subunit CDK-activating kinase (CAK) module composed of CCNH/cyclin H, CDK7 and MNAT1 to form the 10-subunit holoenzyme (holo-TFIIH) active in transcription.
Similarity
Belongs to the TFB1 family.
Keywords
Complete proteome
DNA damage
DNA repair
Nucleus
Reference proteome
Repeat
Transcription
Transcription regulation
Feature
chain General transcription factor IIH subunit 1
Uniprot
H9JKN8
A0A2A4JLH2
A0A2H1WVG4
A0A194QDG7
A0A212EJ66
A0A437B9F2
+ More
A0A067RJ16 A0A026W810 A0A2J7QES5 A0A195BTT7 A0A1W4VZG8 A0A195CED4 A0A034W020 F4WXP5 E2A1N1 A0A0K8WCI6 A0A151JUN4 A0A158N948 A0A0L0CK24 A0A151J7N8 W8C0P6 A0A2A3ERR8 B3NR56 A0A1A9VQA3 A0A1I8MTB2 B4P746 A0A1A9ZWZ4 A0A1B0G4A4 A0A0A1WS92 A0A1W4XGD9 A0A1L8DH93 A0A1A9YLR7 B4QFG2 A0A1A9WF35 A0A151WUY2 A0A1B6FGW6 A0A336M7N3 A0A3B0J1F4 B4HRB5 A0A0C9QVC9 A0A1B6CCQ5 A0A1I8NR24 E2C9K0 A0A0B4K726 Q960E8 A0A023EUH3 B4GAL8 Q291K0 A0A088AUQ9 A0A1B0C799 A0A0L7QL10 B3MDZ8 A0A1Y1M0H5 Q16L69 E0VCF0 B4J482 D2A3F8 A0A1S4FWY9 A0A1Q3F827 T1GW76 A0A232FLB0 B4LQ94 A0A0A9W7P0 A0A182JP18 A0A0J7L207 B4KNX5 A0A182MM03 A0A182T338 B4MQ83 A0A224XA42 A0A023F188 A0A0K8TRP6 U5EYR7 A0A1B0CWD0 A0A182WDP2 T1JKT1 A0A0A9WG22 A0A1B6LDR1 A0A1Y1M5I6 A0A1J1IN71 A0A087UJM6 A0A2R5LCA9 A0A2P6KCA8 R7VIH6 A0A0L8H913 A0A210R191 K1RJ65 A0A0K8SF12 F7BVJ4 A0A444UXK3 A0A2I0UT29 H3ASK4 V8P5L9 W5UJ23 A0A1W7RCQ6 A0A2Y9GUR5 A0A099ZK61 A0A2I0MHP1
A0A067RJ16 A0A026W810 A0A2J7QES5 A0A195BTT7 A0A1W4VZG8 A0A195CED4 A0A034W020 F4WXP5 E2A1N1 A0A0K8WCI6 A0A151JUN4 A0A158N948 A0A0L0CK24 A0A151J7N8 W8C0P6 A0A2A3ERR8 B3NR56 A0A1A9VQA3 A0A1I8MTB2 B4P746 A0A1A9ZWZ4 A0A1B0G4A4 A0A0A1WS92 A0A1W4XGD9 A0A1L8DH93 A0A1A9YLR7 B4QFG2 A0A1A9WF35 A0A151WUY2 A0A1B6FGW6 A0A336M7N3 A0A3B0J1F4 B4HRB5 A0A0C9QVC9 A0A1B6CCQ5 A0A1I8NR24 E2C9K0 A0A0B4K726 Q960E8 A0A023EUH3 B4GAL8 Q291K0 A0A088AUQ9 A0A1B0C799 A0A0L7QL10 B3MDZ8 A0A1Y1M0H5 Q16L69 E0VCF0 B4J482 D2A3F8 A0A1S4FWY9 A0A1Q3F827 T1GW76 A0A232FLB0 B4LQ94 A0A0A9W7P0 A0A182JP18 A0A0J7L207 B4KNX5 A0A182MM03 A0A182T338 B4MQ83 A0A224XA42 A0A023F188 A0A0K8TRP6 U5EYR7 A0A1B0CWD0 A0A182WDP2 T1JKT1 A0A0A9WG22 A0A1B6LDR1 A0A1Y1M5I6 A0A1J1IN71 A0A087UJM6 A0A2R5LCA9 A0A2P6KCA8 R7VIH6 A0A0L8H913 A0A210R191 K1RJ65 A0A0K8SF12 F7BVJ4 A0A444UXK3 A0A2I0UT29 H3ASK4 V8P5L9 W5UJ23 A0A1W7RCQ6 A0A2Y9GUR5 A0A099ZK61 A0A2I0MHP1
Pubmed
19121390
26354079
22118469
24845553
24508170
30249741
+ More
25348373 21719571 20798317 21347285 26108605 24495485 17994087 18057021 25315136 17550304 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 24945155 26483478 15632085 23185243 28004739 17510324 20566863 18362917 19820115 28648823 25401762 26823975 25474469 26369729 23254933 28812685 22992520 17495919 9215903 24297900 23127152 26358130 23371554
25348373 21719571 20798317 21347285 26108605 24495485 17994087 18057021 25315136 17550304 25830018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 24945155 26483478 15632085 23185243 28004739 17510324 20566863 18362917 19820115 28648823 25401762 26823975 25474469 26369729 23254933 28812685 22992520 17495919 9215903 24297900 23127152 26358130 23371554
EMBL
BABH01024909
BABH01024910
NWSH01001172
PCG72260.1
ODYU01011304
SOQ56956.1
+ More
KQ459167 KPJ03502.1 AGBW02014515 OWR41542.1 RSAL01000112 RVE47021.1 KK852655 KDR19295.1 KK107372 QOIP01000004 EZA51786.1 RLU24191.1 NEVH01015305 PNF27094.1 KQ976405 KYM91849.1 KQ977935 KYM98591.1 GAKP01011816 GAKP01011815 GAKP01011814 JAC47138.1 GL888426 EGI61045.1 GL435766 EFN72704.1 GDHF01022889 GDHF01019946 GDHF01003448 JAI29425.1 JAI32368.1 JAI48866.1 KQ981738 KYN36523.1 ADTU01009102 JRES01000378 KNC31824.1 KQ979662 KYN19889.1 GAMC01003902 JAC02654.1 KZ288192 PBC34204.1 CH954179 EDV56045.1 KQS62623.1 CM000158 EDW91011.1 KRJ99569.1 CCAG010004889 GBXI01017288 GBXI01012353 GBXI01007734 JAC97003.1 JAD01939.1 JAD06558.1 GFDF01008329 JAV05755.1 CM000362 CM002911 EDX07072.1 KMY93749.1 KQ982730 KYQ51511.1 GECZ01020328 JAS49441.1 UFQT01000226 SSX22048.1 OUUW01000001 SPP74735.1 CH480816 EDW47844.1 GBYB01007681 GBYB01007684 JAG77448.1 JAG77451.1 GEDC01026051 JAS11247.1 GL453868 EFN75339.1 AE013599 AFH08075.1 AY052097 JXUM01047993 JXUM01047994 GAPW01000543 KQ561542 JAC13055.1 KXJ78206.1 CH479181 EDW31970.1 CM000071 EAL25112.1 KRT01850.1 KRT01851.1 KRT01852.1 JXJN01027938 KQ414934 KOC59280.1 CH902619 EDV37543.1 GEZM01044079 JAV78518.1 CH477919 EAT35058.1 DS235053 EEB11056.1 CH916367 EDW02688.1 KQ971338 EFA02310.1 GFDL01011358 JAV23687.1 CAQQ02153970 CAQQ02153971 NNAY01000051 OXU31531.1 CH940648 EDW61379.1 KRF79969.1 GBHO01039796 GBHO01039795 GDHC01015637 GDHC01007245 GDHC01007205 GDHC01002562 JAG03808.1 JAG03809.1 JAQ02992.1 JAQ11384.1 JAQ11424.1 JAQ16067.1 LBMM01001258 KMQ96628.1 CH933808 EDW09020.1 AXCM01001638 CH963849 EDW74272.1 GFTR01007210 JAW09216.1 GBBI01003669 JAC15043.1 GDAI01000782 JAI16821.1 GANO01000118 JAB59753.1 AJWK01032239 JH431663 GBHO01039794 JAG03810.1 GEBQ01018146 JAT21831.1 GEZM01044080 JAV78517.1 CVRI01000056 CRL01679.1 KK120124 KFM77565.1 GGLE01003024 MBY07150.1 MWRG01016209 PRD23968.1 AMQN01016314 KB291938 ELU18424.1 KQ418812 KOF85771.1 NEDP02000890 OWF54756.1 JH817736 EKC34301.1 GBRD01014137 GBRD01014136 JAG51689.1 SCEB01005433 RXM92923.1 KZ505641 PKU49180.1 AFYH01025600 AFYH01025601 AZIM01000731 ETE69615.1 JT415436 AHH41690.1 GDAY02002551 JAV48916.1 KL895608 KGL82809.1 AKCR02000011 PKK29192.1
KQ459167 KPJ03502.1 AGBW02014515 OWR41542.1 RSAL01000112 RVE47021.1 KK852655 KDR19295.1 KK107372 QOIP01000004 EZA51786.1 RLU24191.1 NEVH01015305 PNF27094.1 KQ976405 KYM91849.1 KQ977935 KYM98591.1 GAKP01011816 GAKP01011815 GAKP01011814 JAC47138.1 GL888426 EGI61045.1 GL435766 EFN72704.1 GDHF01022889 GDHF01019946 GDHF01003448 JAI29425.1 JAI32368.1 JAI48866.1 KQ981738 KYN36523.1 ADTU01009102 JRES01000378 KNC31824.1 KQ979662 KYN19889.1 GAMC01003902 JAC02654.1 KZ288192 PBC34204.1 CH954179 EDV56045.1 KQS62623.1 CM000158 EDW91011.1 KRJ99569.1 CCAG010004889 GBXI01017288 GBXI01012353 GBXI01007734 JAC97003.1 JAD01939.1 JAD06558.1 GFDF01008329 JAV05755.1 CM000362 CM002911 EDX07072.1 KMY93749.1 KQ982730 KYQ51511.1 GECZ01020328 JAS49441.1 UFQT01000226 SSX22048.1 OUUW01000001 SPP74735.1 CH480816 EDW47844.1 GBYB01007681 GBYB01007684 JAG77448.1 JAG77451.1 GEDC01026051 JAS11247.1 GL453868 EFN75339.1 AE013599 AFH08075.1 AY052097 JXUM01047993 JXUM01047994 GAPW01000543 KQ561542 JAC13055.1 KXJ78206.1 CH479181 EDW31970.1 CM000071 EAL25112.1 KRT01850.1 KRT01851.1 KRT01852.1 JXJN01027938 KQ414934 KOC59280.1 CH902619 EDV37543.1 GEZM01044079 JAV78518.1 CH477919 EAT35058.1 DS235053 EEB11056.1 CH916367 EDW02688.1 KQ971338 EFA02310.1 GFDL01011358 JAV23687.1 CAQQ02153970 CAQQ02153971 NNAY01000051 OXU31531.1 CH940648 EDW61379.1 KRF79969.1 GBHO01039796 GBHO01039795 GDHC01015637 GDHC01007245 GDHC01007205 GDHC01002562 JAG03808.1 JAG03809.1 JAQ02992.1 JAQ11384.1 JAQ11424.1 JAQ16067.1 LBMM01001258 KMQ96628.1 CH933808 EDW09020.1 AXCM01001638 CH963849 EDW74272.1 GFTR01007210 JAW09216.1 GBBI01003669 JAC15043.1 GDAI01000782 JAI16821.1 GANO01000118 JAB59753.1 AJWK01032239 JH431663 GBHO01039794 JAG03810.1 GEBQ01018146 JAT21831.1 GEZM01044080 JAV78517.1 CVRI01000056 CRL01679.1 KK120124 KFM77565.1 GGLE01003024 MBY07150.1 MWRG01016209 PRD23968.1 AMQN01016314 KB291938 ELU18424.1 KQ418812 KOF85771.1 NEDP02000890 OWF54756.1 JH817736 EKC34301.1 GBRD01014137 GBRD01014136 JAG51689.1 SCEB01005433 RXM92923.1 KZ505641 PKU49180.1 AFYH01025600 AFYH01025601 AZIM01000731 ETE69615.1 JT415436 AHH41690.1 GDAY02002551 JAV48916.1 KL895608 KGL82809.1 AKCR02000011 PKK29192.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000027135
+ More
UP000053097 UP000279307 UP000235965 UP000078540 UP000192221 UP000078542 UP000007755 UP000000311 UP000078541 UP000005205 UP000037069 UP000078492 UP000242457 UP000008711 UP000078200 UP000095301 UP000002282 UP000092445 UP000092444 UP000192223 UP000092443 UP000000304 UP000091820 UP000075809 UP000268350 UP000001292 UP000095300 UP000008237 UP000000803 UP000069940 UP000249989 UP000008744 UP000001819 UP000005203 UP000092460 UP000053825 UP000007801 UP000008820 UP000009046 UP000001070 UP000007266 UP000015102 UP000215335 UP000008792 UP000075881 UP000036403 UP000009192 UP000075883 UP000075901 UP000007798 UP000092461 UP000075920 UP000183832 UP000054359 UP000014760 UP000053454 UP000242188 UP000005408 UP000002280 UP000008672 UP000221080 UP000248481 UP000053641 UP000053872
UP000053097 UP000279307 UP000235965 UP000078540 UP000192221 UP000078542 UP000007755 UP000000311 UP000078541 UP000005205 UP000037069 UP000078492 UP000242457 UP000008711 UP000078200 UP000095301 UP000002282 UP000092445 UP000092444 UP000192223 UP000092443 UP000000304 UP000091820 UP000075809 UP000268350 UP000001292 UP000095300 UP000008237 UP000000803 UP000069940 UP000249989 UP000008744 UP000001819 UP000005203 UP000092460 UP000053825 UP000007801 UP000008820 UP000009046 UP000001070 UP000007266 UP000015102 UP000215335 UP000008792 UP000075881 UP000036403 UP000009192 UP000075883 UP000075901 UP000007798 UP000092461 UP000075920 UP000183832 UP000054359 UP000014760 UP000053454 UP000242188 UP000005408 UP000002280 UP000008672 UP000221080 UP000248481 UP000053641 UP000053872
Interpro
SUPFAM
SSF54495
SSF54495
Gene 3D
ProteinModelPortal
H9JKN8
A0A2A4JLH2
A0A2H1WVG4
A0A194QDG7
A0A212EJ66
A0A437B9F2
+ More
A0A067RJ16 A0A026W810 A0A2J7QES5 A0A195BTT7 A0A1W4VZG8 A0A195CED4 A0A034W020 F4WXP5 E2A1N1 A0A0K8WCI6 A0A151JUN4 A0A158N948 A0A0L0CK24 A0A151J7N8 W8C0P6 A0A2A3ERR8 B3NR56 A0A1A9VQA3 A0A1I8MTB2 B4P746 A0A1A9ZWZ4 A0A1B0G4A4 A0A0A1WS92 A0A1W4XGD9 A0A1L8DH93 A0A1A9YLR7 B4QFG2 A0A1A9WF35 A0A151WUY2 A0A1B6FGW6 A0A336M7N3 A0A3B0J1F4 B4HRB5 A0A0C9QVC9 A0A1B6CCQ5 A0A1I8NR24 E2C9K0 A0A0B4K726 Q960E8 A0A023EUH3 B4GAL8 Q291K0 A0A088AUQ9 A0A1B0C799 A0A0L7QL10 B3MDZ8 A0A1Y1M0H5 Q16L69 E0VCF0 B4J482 D2A3F8 A0A1S4FWY9 A0A1Q3F827 T1GW76 A0A232FLB0 B4LQ94 A0A0A9W7P0 A0A182JP18 A0A0J7L207 B4KNX5 A0A182MM03 A0A182T338 B4MQ83 A0A224XA42 A0A023F188 A0A0K8TRP6 U5EYR7 A0A1B0CWD0 A0A182WDP2 T1JKT1 A0A0A9WG22 A0A1B6LDR1 A0A1Y1M5I6 A0A1J1IN71 A0A087UJM6 A0A2R5LCA9 A0A2P6KCA8 R7VIH6 A0A0L8H913 A0A210R191 K1RJ65 A0A0K8SF12 F7BVJ4 A0A444UXK3 A0A2I0UT29 H3ASK4 V8P5L9 W5UJ23 A0A1W7RCQ6 A0A2Y9GUR5 A0A099ZK61 A0A2I0MHP1
A0A067RJ16 A0A026W810 A0A2J7QES5 A0A195BTT7 A0A1W4VZG8 A0A195CED4 A0A034W020 F4WXP5 E2A1N1 A0A0K8WCI6 A0A151JUN4 A0A158N948 A0A0L0CK24 A0A151J7N8 W8C0P6 A0A2A3ERR8 B3NR56 A0A1A9VQA3 A0A1I8MTB2 B4P746 A0A1A9ZWZ4 A0A1B0G4A4 A0A0A1WS92 A0A1W4XGD9 A0A1L8DH93 A0A1A9YLR7 B4QFG2 A0A1A9WF35 A0A151WUY2 A0A1B6FGW6 A0A336M7N3 A0A3B0J1F4 B4HRB5 A0A0C9QVC9 A0A1B6CCQ5 A0A1I8NR24 E2C9K0 A0A0B4K726 Q960E8 A0A023EUH3 B4GAL8 Q291K0 A0A088AUQ9 A0A1B0C799 A0A0L7QL10 B3MDZ8 A0A1Y1M0H5 Q16L69 E0VCF0 B4J482 D2A3F8 A0A1S4FWY9 A0A1Q3F827 T1GW76 A0A232FLB0 B4LQ94 A0A0A9W7P0 A0A182JP18 A0A0J7L207 B4KNX5 A0A182MM03 A0A182T338 B4MQ83 A0A224XA42 A0A023F188 A0A0K8TRP6 U5EYR7 A0A1B0CWD0 A0A182WDP2 T1JKT1 A0A0A9WG22 A0A1B6LDR1 A0A1Y1M5I6 A0A1J1IN71 A0A087UJM6 A0A2R5LCA9 A0A2P6KCA8 R7VIH6 A0A0L8H913 A0A210R191 K1RJ65 A0A0K8SF12 F7BVJ4 A0A444UXK3 A0A2I0UT29 H3ASK4 V8P5L9 W5UJ23 A0A1W7RCQ6 A0A2Y9GUR5 A0A099ZK61 A0A2I0MHP1
PDB
6O9M
E-value=2.20066e-104,
Score=970
Ontologies
PATHWAY
GO
PANTHER
Topology
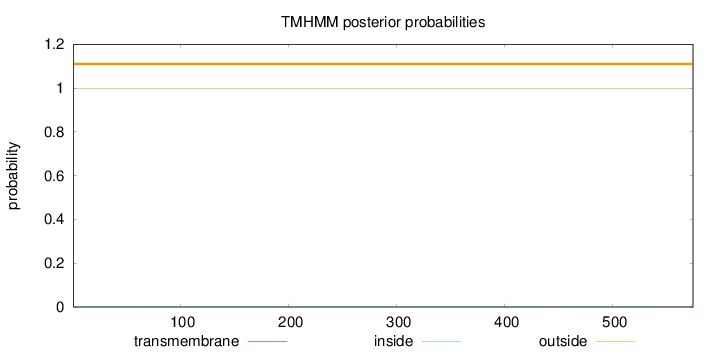
Subcellular location
Nucleus
Length:
574
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00868
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00080
outside
1 - 574
Population Genetic Test Statistics
Pi
151.846498
Theta
136.687346
Tajima's D
0.345441
CLR
2.000372
CSRT
0.474476276186191
Interpretation
Uncertain
