Gene
KWMTBOMO03829 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010088
Annotation
PREDICTED:_AFG3-like_protein_2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.349
Sequence
CDS
ATGTTCTCGCCTTCTAGCAGCAGCGGAGGTGGAAGTGGAGGCCGCGGCGGCTATGAAGGCCAAGATAAAGAGAAGTGGATGATGTTTGGAGCAATTGGTATTGTGACATTGCTTGCTTCAGTTGCGTATTTTGAATTAAGATACAGAGAGATAAGTTGGAGAGATTTTGTCAATTTGTATTTAAACAAAGGTGTGGTCGAGAAACTAGAAGTTATTAACAAAAAATGGGTCCGAGTAAAACTTCAAGGGAATGCATTAGAAGGAAAGGTTATTTGGTTTGCAATAGGTAGTGTCGATTCATTCGAACGTAATCTAGAAAATGCACAAATTGAAATGAGCATTGATCCTCCTAATTTTGTACCAGTTATTTACAAAACTGAAGTAGAGGCTGCCAGTCTGACGGGAATGCTTCCTACTCTACTAATAATTGGGTTTTTAATATACATGATGAGAAGATCTGCAGACATGATGGGACGCGGTGGCCGTCGTGGAGGAGGCTTGTTTGGAGGAGTGATGGAGTCAACAGCTAAATTAATAAATCCAACAGACATTGGAGTAAAATTCCAAGATGTTGCTGGTTGTGAAGAAGCAAAGATAGAAATTATGGAATTTGTCAACTTTCTTAAAAACCCTCAGCAATATATAGATTTGGGAGCTAAAATTCCCAAAGGTGCACTATTAACAGGTCCTCCCGGTACTGGTAAAACTTTACTCGCAAAAGCAACTGCAGGAGAAGCTAATGTACCATTTATCACTGTCTCTGGTTCAGAATTCTTAGAGATGTTTGTAGGTGTTGGTCCTTCCAGAGTAAGAGATATGTTTTCTATGGCACGTAAACATGCTCCATGCATACTGTTTATTGATGAAATTGATGCTGTAGGTAGAAAAAGAGGAGGTCGTAGTTTTGGTGGTCATTCAGAACAAGAAAACACTTTGAATCAACTGTTGGTTGAAATGGATGGATTTAATACAACTACAAATGTTGTGGTACTTGCAGCTACAAATCGAGTAGATATATTAGATAAGGCATTGTTGAGACCTGGTAGATTTGATAGACAGATATTTGTCCCAGCACCAGACATTAAGGGCAGGGCCTCAATTTTCAAGGTGCATTTGACTCCTCTCAAGACAACGCTGAATAAGGAAAATTTAGCAAGAAAAATGGCTGCATTGACTCCAGGTTTCACCGGCGCGGATATTGCTAACGTGTGCAACGAGGCGGCACTAATTGCCGCTCGCGAATTAGCCAACGATATTACAATGAAAAATTTCGAACAAGCCATCGAAAGAGTCGTCGCCGGTATGGAGAAAAAATCTAATGTTCTCCAACCAGACGAGAGGAAGATTGTAGCGTATCATGAGGCCGGCCATGCAGTTGCAGGTTGGTTCCTACAACATGCAGATCCCCTTTTAAAAGTGTCTATTATACCTAGAGGAAAAGGTCTTGGCTACGCGCAATATTTACCCAAAGAACAGTATTTATACAGCAAGGAACAACTATTTGATAGAATGTGCATGACACTAGGGGGTAGAGTAAGTGAAGAGATATTCTTCGGAAGAATAACCACAGGGGCTCAGGATGACTTAAAAAAGATAACACAAAGCGCTTATGCGCAGATTGTACACTATGGTATGAACGCTAAAGTCGGAAATGTGTCTTTCGAGATGCCACAGCCCGGTGAAATGGTGATTGACAAACCATACTCTGAAAAGACCGCAGAACTCATCGATTCAGAAGTCAGAGACCTGATTAACAATGCGCACAAACACACAACGGATTTATTAACAAAACACAAACCTAACATCGAAAAGGTTGCAGAGAGATTACTGAAACAGGAGATTTTGAGTCGCGATGATATGATTGAACTGTTAGGGCCTAGACCGTTCCCCGAGAAGAGTACTTATGAAGAGTTCGTTGAAGGCACAGGATCATTAGATGAAGACACGACGCTACCAGAAGGCCTCAAGGATTGGAATAAGGAGAAACAGCCCACAACACCACCGCCGGAAACCAGCCCATCAAGCCCCACAGCCAGTAAAAAATAA
Protein
MFSPSSSSGGGSGGRGGYEGQDKEKWMMFGAIGIVTLLASVAYFELRYREISWRDFVNLYLNKGVVEKLEVINKKWVRVKLQGNALEGKVIWFAIGSVDSFERNLENAQIEMSIDPPNFVPVIYKTEVEAASLTGMLPTLLIIGFLIYMMRRSADMMGRGGRRGGGLFGGVMESTAKLINPTDIGVKFQDVAGCEEAKIEIMEFVNFLKNPQQYIDLGAKIPKGALLTGPPGTGKTLLAKATAGEANVPFITVSGSEFLEMFVGVGPSRVRDMFSMARKHAPCILFIDEIDAVGRKRGGRSFGGHSEQENTLNQLLVEMDGFNTTTNVVVLAATNRVDILDKALLRPGRFDRQIFVPAPDIKGRASIFKVHLTPLKTTLNKENLARKMAALTPGFTGADIANVCNEAALIAARELANDITMKNFEQAIERVVAGMEKKSNVLQPDERKIVAYHEAGHAVAGWFLQHADPLLKVSIIPRGKGLGYAQYLPKEQYLYSKEQLFDRMCMTLGGRVSEEIFFGRITTGAQDDLKKITQSAYAQIVHYGMNAKVGNVSFEMPQPGEMVIDKPYSEKTAELIDSEVRDLINNAHKHTTDLLTKHKPNIEKVAERLLKQEILSRDDMIELLGPRPFPEKSTYEEFVEGTGSLDEDTTLPEGLKDWNKEKQPTTPPPETSPSSPTASKK
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JKN7
A0A194QEX5
A0A194QZK1
A0A0L7LM74
A0A2A4JLH3
A0A2H1WV73
+ More
A0A1B6D736 A0A067QGZ3 A0A2R7VV90 A0A2J7PHK7 A0A0P4VNS7 T1HR87 A0A069DXI0 A0A0V0G4E0 A0A224XBN9 D6WI24 A0A023F5C5 A0A1W4XFS0 A0A0K8TLD1 A0A1S4ETA6 U5EYX2 A0A0A9YLQ2 A0A0K8SQB5 W8C7T2 A0A0P5CVC5 A0A0P5LGG8 A0A0P5HNW7 A0A0P5QV32 A0A0P6FUV9 A0A0P4XPH9 A0A0P4XL91 A0A0N8AFI2 A0A0P5ZWZ4 A0A0P4WQV2 A0A0P5XLV7 A0A0P5E339 A0A0A1X802 A0A0P5NL27 A0A0P5HLN8 A0A0P5J9W4 A0A0P5ASG6 A0A0P4WUH2 A0A0P5XLT1 A0A0P6BRQ6 E9G6N8 A0A0P5MNS2 A0A0N8CLA8 A0A0P5B3T1 A0A0P5CJM6 A0A0K8WJT3 A0A0P5ADX4 A0A0P5P212 A0A1J1IK09 A0A0P6G3R5 A0A0P5D3X5 A0A0N8D4A8 A0A034W644 A0A226DFQ1 A0A0K8TXS8 A0A0N8CR39 A0A0P5CS56 A0A0P4WLC9 A0A0P5W0E1 A0A0P5A260 A0A182H2R4 A0A1S4FJU4 A0A0P5U7L4 A0A2M3ZGZ5 A0A2M3Z4U6 A0A0P5ZQL4 A0A2M4BDX4 Q16YE9 A0A0C9QH78 A0A2M4BE63 N6UDT8 E2AAL5 A0A2M4AD25 A0A0P5BVN1 B4PJY1 A0A0L0BVM1 A0A1L8D996 A0A2R5LML1 A0A182FGT8 B3NHX7 A0A131Y2Z3 W5JS15 A0A1B0GI02 Q8T4G5 B4QNF5 Q8IQQ9 A0A1Y1N3C4 A0A1L8D8U0 A0A1E1X2W7 A0A1I8PIZ2 T1IJ28 A0A0K8RNM4 A0A1W4VSD6 A0A1A9WFI9 A0A182J1D4 A0A293N4Y3
A0A1B6D736 A0A067QGZ3 A0A2R7VV90 A0A2J7PHK7 A0A0P4VNS7 T1HR87 A0A069DXI0 A0A0V0G4E0 A0A224XBN9 D6WI24 A0A023F5C5 A0A1W4XFS0 A0A0K8TLD1 A0A1S4ETA6 U5EYX2 A0A0A9YLQ2 A0A0K8SQB5 W8C7T2 A0A0P5CVC5 A0A0P5LGG8 A0A0P5HNW7 A0A0P5QV32 A0A0P6FUV9 A0A0P4XPH9 A0A0P4XL91 A0A0N8AFI2 A0A0P5ZWZ4 A0A0P4WQV2 A0A0P5XLV7 A0A0P5E339 A0A0A1X802 A0A0P5NL27 A0A0P5HLN8 A0A0P5J9W4 A0A0P5ASG6 A0A0P4WUH2 A0A0P5XLT1 A0A0P6BRQ6 E9G6N8 A0A0P5MNS2 A0A0N8CLA8 A0A0P5B3T1 A0A0P5CJM6 A0A0K8WJT3 A0A0P5ADX4 A0A0P5P212 A0A1J1IK09 A0A0P6G3R5 A0A0P5D3X5 A0A0N8D4A8 A0A034W644 A0A226DFQ1 A0A0K8TXS8 A0A0N8CR39 A0A0P5CS56 A0A0P4WLC9 A0A0P5W0E1 A0A0P5A260 A0A182H2R4 A0A1S4FJU4 A0A0P5U7L4 A0A2M3ZGZ5 A0A2M3Z4U6 A0A0P5ZQL4 A0A2M4BDX4 Q16YE9 A0A0C9QH78 A0A2M4BE63 N6UDT8 E2AAL5 A0A2M4AD25 A0A0P5BVN1 B4PJY1 A0A0L0BVM1 A0A1L8D996 A0A2R5LML1 A0A182FGT8 B3NHX7 A0A131Y2Z3 W5JS15 A0A1B0GI02 Q8T4G5 B4QNF5 Q8IQQ9 A0A1Y1N3C4 A0A1L8D8U0 A0A1E1X2W7 A0A1I8PIZ2 T1IJ28 A0A0K8RNM4 A0A1W4VSD6 A0A1A9WFI9 A0A182J1D4 A0A293N4Y3
Pubmed
19121390
26354079
26227816
24845553
27129103
26334808
+ More
18362917 19820115 25474469 26369729 25401762 24495485 25830018 21292972 25348373 26483478 17510324 23537049 20798317 17994087 17550304 26108605 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28004739 28503490
18362917 19820115 25474469 26369729 25401762 24495485 25830018 21292972 25348373 26483478 17510324 23537049 20798317 17994087 17550304 26108605 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 28004739 28503490
EMBL
BABH01024905
KQ459167
KPJ03500.1
KQ460930
KPJ10714.1
JTDY01000656
+ More
KOB76311.1 NWSH01001172 PCG72262.1 ODYU01011304 SOQ56958.1 GEDC01015800 JAS21498.1 KK853424 KDR07700.1 KK854089 PTY11078.1 NEVH01025136 PNF15813.1 GDKW01000249 JAI56346.1 ACPB03025706 GBGD01000502 JAC88387.1 GECL01003217 JAP02907.1 GFTR01008062 JAW08364.1 KQ971334 EEZ99702.1 GBBI01002393 JAC16319.1 GDAI01002878 JAI14725.1 GANO01001763 JAB58108.1 GBHO01011576 JAG32028.1 GBRD01010373 JAG55451.1 GAMC01003429 GAMC01003428 JAC03127.1 GDIP01165030 JAJ58372.1 GDIQ01171503 JAK80222.1 GDIQ01224384 JAK27341.1 GDIQ01118673 JAL33053.1 GDIQ01053855 JAN40882.1 GDIP01238628 JAI84773.1 GDIP01239724 JAI83677.1 GDIP01152481 JAJ70921.1 GDIP01037443 JAM66272.1 GDIP01252012 GDIP01195885 GDIP01080991 GDIP01045099 LRGB01000337 JAI71389.1 KZS19665.1 GDIP01070184 JAM33531.1 GDIP01165774 JAJ57628.1 GBXI01007447 JAD06845.1 GDIQ01177399 GDIQ01148729 GDIQ01140773 JAL10953.1 GDIQ01225590 JAK26135.1 GDIQ01208948 JAK42777.1 GDIP01194680 JAJ28722.1 GDIP01252011 GDIP01195884 GDIP01080990 GDIP01045098 JAI71390.1 GDIP01070237 JAM33478.1 GDIP01011123 JAM92592.1 GL732533 EFX85159.1 GDIQ01155608 JAK96117.1 GDIP01120583 JAL83131.1 GDIP01194679 JAJ28723.1 GDIP01169117 JAJ54285.1 GDHF01027841 GDHF01021403 GDHF01000967 GDHF01000154 JAI24473.1 JAI30911.1 JAI51347.1 JAI52160.1 GDIP01203575 JAJ19827.1 GDIQ01155609 JAK96116.1 CVRI01000054 CRL00090.1 GDIQ01038864 JAN55873.1 GDIP01165773 JAJ57629.1 GDIP01070183 JAM33532.1 GAKP01009730 GAKP01009729 JAC49223.1 LNIX01000021 OXA43808.1 GDHF01033218 JAI19096.1 GDIP01107135 JAL96579.1 GDIP01168314 GDIP01167610 JAJ55792.1 GDIP01254664 JAI68737.1 GDIP01107136 JAL96578.1 GDIP01217422 JAJ05980.1 JXUM01024012 JXUM01024013 JXUM01024014 JXUM01024015 KQ560677 KXJ81315.1 GDIP01118481 JAL85233.1 GGFM01007086 MBW27837.1 GGFM01002789 MBW23540.1 GDIP01039701 JAM64014.1 GGFJ01002104 MBW51245.1 CH477517 EAT39655.1 GBYB01002848 GBYB01002849 JAG72615.1 JAG72616.1 GGFJ01002110 MBW51251.1 APGK01026583 APGK01026584 KB740635 ENN79870.1 GL438129 EFN69522.1 GGFK01005362 MBW38683.1 GDIP01179280 JAJ44122.1 CM000159 EDW94750.1 JRES01001371 KNC23284.1 GFDF01011210 JAV02874.1 GGLE01006647 MBY10773.1 CH954178 EDV51992.1 GEFM01002918 JAP72878.1 ADMH02000528 ETN66093.1 AJWK01010543 AE014296 AY084199 AAF49365.2 AAL89937.1 CM000363 CM002912 EDX10825.1 KMZ00202.1 AAN11704.1 GEZM01013648 GEZM01013647 JAV92404.1 GFDF01011211 JAV02873.1 GFAC01005802 JAT93386.1 JH430221 GADI01001619 JAA72189.1 GFWV01022742 MAA47469.1
KOB76311.1 NWSH01001172 PCG72262.1 ODYU01011304 SOQ56958.1 GEDC01015800 JAS21498.1 KK853424 KDR07700.1 KK854089 PTY11078.1 NEVH01025136 PNF15813.1 GDKW01000249 JAI56346.1 ACPB03025706 GBGD01000502 JAC88387.1 GECL01003217 JAP02907.1 GFTR01008062 JAW08364.1 KQ971334 EEZ99702.1 GBBI01002393 JAC16319.1 GDAI01002878 JAI14725.1 GANO01001763 JAB58108.1 GBHO01011576 JAG32028.1 GBRD01010373 JAG55451.1 GAMC01003429 GAMC01003428 JAC03127.1 GDIP01165030 JAJ58372.1 GDIQ01171503 JAK80222.1 GDIQ01224384 JAK27341.1 GDIQ01118673 JAL33053.1 GDIQ01053855 JAN40882.1 GDIP01238628 JAI84773.1 GDIP01239724 JAI83677.1 GDIP01152481 JAJ70921.1 GDIP01037443 JAM66272.1 GDIP01252012 GDIP01195885 GDIP01080991 GDIP01045099 LRGB01000337 JAI71389.1 KZS19665.1 GDIP01070184 JAM33531.1 GDIP01165774 JAJ57628.1 GBXI01007447 JAD06845.1 GDIQ01177399 GDIQ01148729 GDIQ01140773 JAL10953.1 GDIQ01225590 JAK26135.1 GDIQ01208948 JAK42777.1 GDIP01194680 JAJ28722.1 GDIP01252011 GDIP01195884 GDIP01080990 GDIP01045098 JAI71390.1 GDIP01070237 JAM33478.1 GDIP01011123 JAM92592.1 GL732533 EFX85159.1 GDIQ01155608 JAK96117.1 GDIP01120583 JAL83131.1 GDIP01194679 JAJ28723.1 GDIP01169117 JAJ54285.1 GDHF01027841 GDHF01021403 GDHF01000967 GDHF01000154 JAI24473.1 JAI30911.1 JAI51347.1 JAI52160.1 GDIP01203575 JAJ19827.1 GDIQ01155609 JAK96116.1 CVRI01000054 CRL00090.1 GDIQ01038864 JAN55873.1 GDIP01165773 JAJ57629.1 GDIP01070183 JAM33532.1 GAKP01009730 GAKP01009729 JAC49223.1 LNIX01000021 OXA43808.1 GDHF01033218 JAI19096.1 GDIP01107135 JAL96579.1 GDIP01168314 GDIP01167610 JAJ55792.1 GDIP01254664 JAI68737.1 GDIP01107136 JAL96578.1 GDIP01217422 JAJ05980.1 JXUM01024012 JXUM01024013 JXUM01024014 JXUM01024015 KQ560677 KXJ81315.1 GDIP01118481 JAL85233.1 GGFM01007086 MBW27837.1 GGFM01002789 MBW23540.1 GDIP01039701 JAM64014.1 GGFJ01002104 MBW51245.1 CH477517 EAT39655.1 GBYB01002848 GBYB01002849 JAG72615.1 JAG72616.1 GGFJ01002110 MBW51251.1 APGK01026583 APGK01026584 KB740635 ENN79870.1 GL438129 EFN69522.1 GGFK01005362 MBW38683.1 GDIP01179280 JAJ44122.1 CM000159 EDW94750.1 JRES01001371 KNC23284.1 GFDF01011210 JAV02874.1 GGLE01006647 MBY10773.1 CH954178 EDV51992.1 GEFM01002918 JAP72878.1 ADMH02000528 ETN66093.1 AJWK01010543 AE014296 AY084199 AAF49365.2 AAL89937.1 CM000363 CM002912 EDX10825.1 KMZ00202.1 AAN11704.1 GEZM01013648 GEZM01013647 JAV92404.1 GFDF01011211 JAV02873.1 GFAC01005802 JAT93386.1 JH430221 GADI01001619 JAA72189.1 GFWV01022742 MAA47469.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000218220
UP000027135
+ More
UP000235965 UP000015103 UP000007266 UP000192223 UP000079169 UP000076858 UP000000305 UP000183832 UP000198287 UP000069940 UP000249989 UP000008820 UP000019118 UP000000311 UP000002282 UP000037069 UP000069272 UP000008711 UP000000673 UP000092461 UP000000803 UP000000304 UP000095300 UP000192221 UP000091820 UP000075880
UP000235965 UP000015103 UP000007266 UP000192223 UP000079169 UP000076858 UP000000305 UP000183832 UP000198287 UP000069940 UP000249989 UP000008820 UP000019118 UP000000311 UP000002282 UP000037069 UP000069272 UP000008711 UP000000673 UP000092461 UP000000803 UP000000304 UP000095300 UP000192221 UP000091820 UP000075880
Interpro
Gene 3D
ProteinModelPortal
H9JKN7
A0A194QEX5
A0A194QZK1
A0A0L7LM74
A0A2A4JLH3
A0A2H1WV73
+ More
A0A1B6D736 A0A067QGZ3 A0A2R7VV90 A0A2J7PHK7 A0A0P4VNS7 T1HR87 A0A069DXI0 A0A0V0G4E0 A0A224XBN9 D6WI24 A0A023F5C5 A0A1W4XFS0 A0A0K8TLD1 A0A1S4ETA6 U5EYX2 A0A0A9YLQ2 A0A0K8SQB5 W8C7T2 A0A0P5CVC5 A0A0P5LGG8 A0A0P5HNW7 A0A0P5QV32 A0A0P6FUV9 A0A0P4XPH9 A0A0P4XL91 A0A0N8AFI2 A0A0P5ZWZ4 A0A0P4WQV2 A0A0P5XLV7 A0A0P5E339 A0A0A1X802 A0A0P5NL27 A0A0P5HLN8 A0A0P5J9W4 A0A0P5ASG6 A0A0P4WUH2 A0A0P5XLT1 A0A0P6BRQ6 E9G6N8 A0A0P5MNS2 A0A0N8CLA8 A0A0P5B3T1 A0A0P5CJM6 A0A0K8WJT3 A0A0P5ADX4 A0A0P5P212 A0A1J1IK09 A0A0P6G3R5 A0A0P5D3X5 A0A0N8D4A8 A0A034W644 A0A226DFQ1 A0A0K8TXS8 A0A0N8CR39 A0A0P5CS56 A0A0P4WLC9 A0A0P5W0E1 A0A0P5A260 A0A182H2R4 A0A1S4FJU4 A0A0P5U7L4 A0A2M3ZGZ5 A0A2M3Z4U6 A0A0P5ZQL4 A0A2M4BDX4 Q16YE9 A0A0C9QH78 A0A2M4BE63 N6UDT8 E2AAL5 A0A2M4AD25 A0A0P5BVN1 B4PJY1 A0A0L0BVM1 A0A1L8D996 A0A2R5LML1 A0A182FGT8 B3NHX7 A0A131Y2Z3 W5JS15 A0A1B0GI02 Q8T4G5 B4QNF5 Q8IQQ9 A0A1Y1N3C4 A0A1L8D8U0 A0A1E1X2W7 A0A1I8PIZ2 T1IJ28 A0A0K8RNM4 A0A1W4VSD6 A0A1A9WFI9 A0A182J1D4 A0A293N4Y3
A0A1B6D736 A0A067QGZ3 A0A2R7VV90 A0A2J7PHK7 A0A0P4VNS7 T1HR87 A0A069DXI0 A0A0V0G4E0 A0A224XBN9 D6WI24 A0A023F5C5 A0A1W4XFS0 A0A0K8TLD1 A0A1S4ETA6 U5EYX2 A0A0A9YLQ2 A0A0K8SQB5 W8C7T2 A0A0P5CVC5 A0A0P5LGG8 A0A0P5HNW7 A0A0P5QV32 A0A0P6FUV9 A0A0P4XPH9 A0A0P4XL91 A0A0N8AFI2 A0A0P5ZWZ4 A0A0P4WQV2 A0A0P5XLV7 A0A0P5E339 A0A0A1X802 A0A0P5NL27 A0A0P5HLN8 A0A0P5J9W4 A0A0P5ASG6 A0A0P4WUH2 A0A0P5XLT1 A0A0P6BRQ6 E9G6N8 A0A0P5MNS2 A0A0N8CLA8 A0A0P5B3T1 A0A0P5CJM6 A0A0K8WJT3 A0A0P5ADX4 A0A0P5P212 A0A1J1IK09 A0A0P6G3R5 A0A0P5D3X5 A0A0N8D4A8 A0A034W644 A0A226DFQ1 A0A0K8TXS8 A0A0N8CR39 A0A0P5CS56 A0A0P4WLC9 A0A0P5W0E1 A0A0P5A260 A0A182H2R4 A0A1S4FJU4 A0A0P5U7L4 A0A2M3ZGZ5 A0A2M3Z4U6 A0A0P5ZQL4 A0A2M4BDX4 Q16YE9 A0A0C9QH78 A0A2M4BE63 N6UDT8 E2AAL5 A0A2M4AD25 A0A0P5BVN1 B4PJY1 A0A0L0BVM1 A0A1L8D996 A0A2R5LML1 A0A182FGT8 B3NHX7 A0A131Y2Z3 W5JS15 A0A1B0GI02 Q8T4G5 B4QNF5 Q8IQQ9 A0A1Y1N3C4 A0A1L8D8U0 A0A1E1X2W7 A0A1I8PIZ2 T1IJ28 A0A0K8RNM4 A0A1W4VSD6 A0A1A9WFI9 A0A182J1D4 A0A293N4Y3
PDB
6NYY
E-value=0,
Score=2003
Ontologies
GO
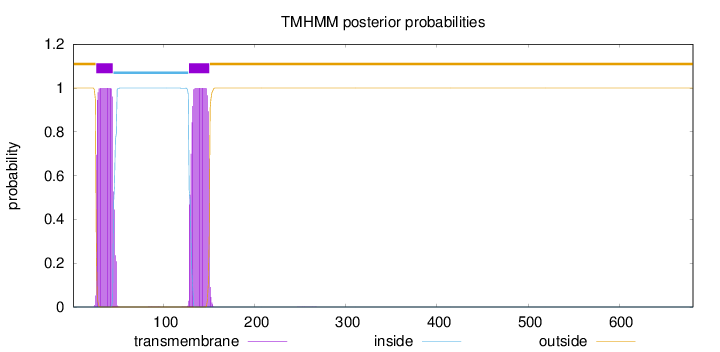
Topology
Length:
681
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.2392500000001
Exp number, first 60 AAs:
20.05883
Total prob of N-in:
0.00019
POSSIBLE N-term signal
sequence
outside
1 - 25
TMhelix
26 - 44
inside
45 - 127
TMhelix
128 - 150
outside
151 - 681
Population Genetic Test Statistics
Pi
224.211946
Theta
206.5491
Tajima's D
0.301478
CLR
359.59914
CSRT
0.451677416129194
Interpretation
Uncertain
