Gene
KWMTBOMO03827
Annotation
_gustatory_receptor_16_[Bombyx_mori]
Full name
Gustatory receptor
Location in the cell
PlasmaMembrane Reliability : 4.204
Sequence
CDS
ATGATGTCCATATACTACACGCTGTTTCAAGTGATGAACAAGCATTTTCAGGAAGTGGCTCCAGTGCTGAGCCTGAGCTACTTCACGATAATTCTGTCCGTGCTTTTGATTTTGGCACCCTCGTTATGCGCCGGAGTGCTACCATGGAAAACTAGACATATGAGACTTATCCTTCTAGAGAAATTGTTTGCAGAAAAAGACAAAAACTCAGCTCGAGAGATTGAGCTGTTCATCAAATATATAGAAGCACGACCCCTTCAGCTTCGAGCCTGCAATTTGGTACCTCTCGATTTCAACCTCCCTGTAATTGTATTAAATTTATGCATTACATATCTGATTGTGATTATACAATTTACGCACTTATTTTAA
Protein
MMSIYYTLFQVMNKHFQEVAPVLSLSYFTIILSVLLILAPSLCAGVLPWKTRHMRLILLEKLFAEKDKNSAREIELFIKYIEARPLQLRACNLVPLDFNLPVIVLNLCITYLIVIIQFTHLF
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Similarity
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Uniprot
B7FF32
B7FF31
B7FF30
B7FF33
A0A3S2TIL7
A0A0L7LBX6
+ More
A0A0L7LBC9 A0A1X9PF35 A0A0L7LB58 A0A0L7KVE4 A0A0L7KTZ0 A0A0L7KU50 A0A437AWF5 A0A0L7K2U9 A0A0L7L2D4 B7FF34 A0A1B3P5S7 A0A1E1W3C9 A0A1E1W4B0 A0A3S2LYE6 A0A212F8B9 A0A2H1W883 A0A2W1B1C4 A0A437B4C8 H9JKN4 A0A2H1WVF0 A0A2H1VJW5 A0A2H1WX21 H9JKN6 A0A2W1BCN5 A0A1L7NV11 B7FF38 A0A194QYU4 B7FF37 A0A1L7NV17 A0A2A4IVK0 A0A2A4JTB6 A0A386H949 A0A2W1B875 A0A2W1B9N9 A0A2H1VG63 A0A2W1BBR6 A0A067RU62 A0A1L7NV12 B7FF36 A0A087ZNS5 A0A2K8GL65 E2AGX1 A0A2A3EQ19 A0A2H1W3R6 A0A2H1VWL2
A0A0L7LBC9 A0A1X9PF35 A0A0L7LB58 A0A0L7KVE4 A0A0L7KTZ0 A0A0L7KU50 A0A437AWF5 A0A0L7K2U9 A0A0L7L2D4 B7FF34 A0A1B3P5S7 A0A1E1W3C9 A0A1E1W4B0 A0A3S2LYE6 A0A212F8B9 A0A2H1W883 A0A2W1B1C4 A0A437B4C8 H9JKN4 A0A2H1WVF0 A0A2H1VJW5 A0A2H1WX21 H9JKN6 A0A2W1BCN5 A0A1L7NV11 B7FF38 A0A194QYU4 B7FF37 A0A1L7NV17 A0A2A4IVK0 A0A2A4JTB6 A0A386H949 A0A2W1B875 A0A2W1B9N9 A0A2H1VG63 A0A2W1BBR6 A0A067RU62 A0A1L7NV12 B7FF36 A0A087ZNS5 A0A2K8GL65 E2AGX1 A0A2A3EQ19 A0A2H1W3R6 A0A2H1VWL2
EMBL
BK006599
DAA06379.1
BK006598
DAA06378.1
BK006597
DAA06377.1
+ More
LC056060 BK006600 BAW33757.1 DAA06380.1 RSAL01000112 RVE47015.1 JTDY01001857 KOB72706.1 KOB72705.1 KX096213 ARO76487.1 KOB72707.1 JTDY01005391 KOB67056.1 JTDY01005825 KOB66575.1 KOB66574.1 RSAL01000337 RVE42431.1 JTDY01013653 KOB52116.1 JTDY01003401 KOB69617.1 BK006601 DAA06381.1 KX656014 AOG12963.1 GDQN01009663 JAT81391.1 GDQN01009237 JAT81817.1 RVE47016.1 AGBW02009760 OWR49985.1 ODYU01006932 SOQ49223.1 KZ150469 PZC70769.1 RSAL01000164 RVE45342.1 BABH01024875 ODYU01011304 SOQ56962.1 ODYU01002950 SOQ41096.1 SOQ56964.1 BABH01024895 PZC70766.1 BAW33747.1 BK006605 BAW33748.1 BAW33749.1 BAW33750.1 BAW33751.1 BAW33752.1 BAW33753.1 BAW33754.1 BAW33755.1 DAA06385.1 KQ460930 KPJ10708.1 BK006604 DAA06384.1 BAW33756.1 NWSH01006857 PCG63212.1 NWSH01000664 PCG74938.1 MG546665 AYD42288.1 PZC70765.1 PZC70764.1 ODYU01002402 SOQ39838.1 PZC70767.1 KK852415 KDR24350.1 BAW33746.1 BK006603 BAW33745.1 DAA06383.1 KY225525 ARO70537.1 GL439417 EFN67318.1 KZ288204 PBC33326.1 ODYU01006143 SOQ47739.1 ODYU01004719 SOQ44902.1
LC056060 BK006600 BAW33757.1 DAA06380.1 RSAL01000112 RVE47015.1 JTDY01001857 KOB72706.1 KOB72705.1 KX096213 ARO76487.1 KOB72707.1 JTDY01005391 KOB67056.1 JTDY01005825 KOB66575.1 KOB66574.1 RSAL01000337 RVE42431.1 JTDY01013653 KOB52116.1 JTDY01003401 KOB69617.1 BK006601 DAA06381.1 KX656014 AOG12963.1 GDQN01009663 JAT81391.1 GDQN01009237 JAT81817.1 RVE47016.1 AGBW02009760 OWR49985.1 ODYU01006932 SOQ49223.1 KZ150469 PZC70769.1 RSAL01000164 RVE45342.1 BABH01024875 ODYU01011304 SOQ56962.1 ODYU01002950 SOQ41096.1 SOQ56964.1 BABH01024895 PZC70766.1 BAW33747.1 BK006605 BAW33748.1 BAW33749.1 BAW33750.1 BAW33751.1 BAW33752.1 BAW33753.1 BAW33754.1 BAW33755.1 DAA06385.1 KQ460930 KPJ10708.1 BK006604 DAA06384.1 BAW33756.1 NWSH01006857 PCG63212.1 NWSH01000664 PCG74938.1 MG546665 AYD42288.1 PZC70765.1 PZC70764.1 ODYU01002402 SOQ39838.1 PZC70767.1 KK852415 KDR24350.1 BAW33746.1 BK006603 BAW33745.1 DAA06383.1 KY225525 ARO70537.1 GL439417 EFN67318.1 KZ288204 PBC33326.1 ODYU01006143 SOQ47739.1 ODYU01004719 SOQ44902.1
Proteomes
Pfam
PF08395 7tm_7
Interpro
IPR013604
7TM_chemorcpt
ProteinModelPortal
B7FF32
B7FF31
B7FF30
B7FF33
A0A3S2TIL7
A0A0L7LBX6
+ More
A0A0L7LBC9 A0A1X9PF35 A0A0L7LB58 A0A0L7KVE4 A0A0L7KTZ0 A0A0L7KU50 A0A437AWF5 A0A0L7K2U9 A0A0L7L2D4 B7FF34 A0A1B3P5S7 A0A1E1W3C9 A0A1E1W4B0 A0A3S2LYE6 A0A212F8B9 A0A2H1W883 A0A2W1B1C4 A0A437B4C8 H9JKN4 A0A2H1WVF0 A0A2H1VJW5 A0A2H1WX21 H9JKN6 A0A2W1BCN5 A0A1L7NV11 B7FF38 A0A194QYU4 B7FF37 A0A1L7NV17 A0A2A4IVK0 A0A2A4JTB6 A0A386H949 A0A2W1B875 A0A2W1B9N9 A0A2H1VG63 A0A2W1BBR6 A0A067RU62 A0A1L7NV12 B7FF36 A0A087ZNS5 A0A2K8GL65 E2AGX1 A0A2A3EQ19 A0A2H1W3R6 A0A2H1VWL2
A0A0L7LBC9 A0A1X9PF35 A0A0L7LB58 A0A0L7KVE4 A0A0L7KTZ0 A0A0L7KU50 A0A437AWF5 A0A0L7K2U9 A0A0L7L2D4 B7FF34 A0A1B3P5S7 A0A1E1W3C9 A0A1E1W4B0 A0A3S2LYE6 A0A212F8B9 A0A2H1W883 A0A2W1B1C4 A0A437B4C8 H9JKN4 A0A2H1WVF0 A0A2H1VJW5 A0A2H1WX21 H9JKN6 A0A2W1BCN5 A0A1L7NV11 B7FF38 A0A194QYU4 B7FF37 A0A1L7NV17 A0A2A4IVK0 A0A2A4JTB6 A0A386H949 A0A2W1B875 A0A2W1B9N9 A0A2H1VG63 A0A2W1BBR6 A0A067RU62 A0A1L7NV12 B7FF36 A0A087ZNS5 A0A2K8GL65 E2AGX1 A0A2A3EQ19 A0A2H1W3R6 A0A2H1VWL2
Ontologies
KEGG
GO
PANTHER
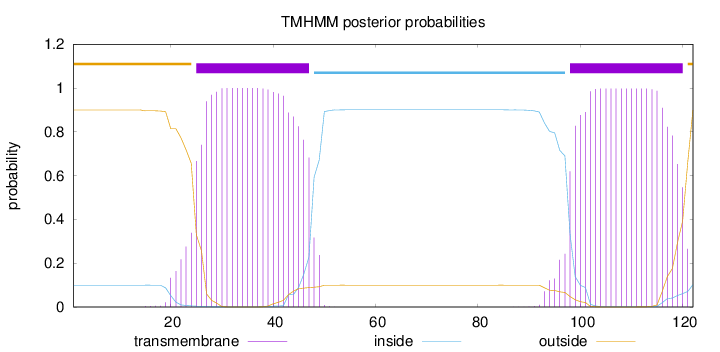
Topology
Subcellular location
Cell membrane
Length:
122
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.89879
Exp number, first 60 AAs:
22.97054
Total prob of N-in:
0.09984
POSSIBLE N-term signal
sequence
outside
1 - 24
TMhelix
25 - 47
inside
48 - 97
TMhelix
98 - 120
outside
121 - 122
Population Genetic Test Statistics
Pi
200.601293
Theta
196.692082
Tajima's D
0.082155
CLR
124.049639
CSRT
0.392880355982201
Interpretation
Uncertain
