Gene
KWMTBOMO03817
Pre Gene Modal
BGIBMGA010008
Annotation
PREDICTED:_uncharacterized_protein_LOC106131468_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.347 Mitochondrial Reliability : 1.052
Sequence
CDS
ATGTATCGTGGCATTCTAGTTTTCTTAACCATTTATCTGACTACTTCTGAAGCTGTTACCAACAGTACAAATTGTGGTTATTTTCAAGCCGACTTTGATTTGATATCTGCCTTAGGGACTTGGCATGTTGTAGCTATTATACCAGAGAAGTTATTTCCGGATAAAGATGTCACTTGTTATAAAATGGAAATCAGCGAAACTGATGAGGCCGGTCTTCGATGGTTGATAAATAGAACCACACATGGACCTCAAAATAAGTCCATAATAGCAGATGTGAAAGGCACGATAGTGAGGCAGAGGTACCATACGGAACATCCCTTTGATGTTTGGTCTAAGTCTACGGCTGGAATCAAAGGATGCTTTCAACAAGTTCTATCCTTGGATTTAGATAAAAACCGAATCCATAAAACCTTAGCTCATGATGCTATGATGCAGTTGCACATAGTGGACTCCAAAGATGGTTCTAGTCCGTTTCTGATGCAAATGCTCTGGGGGAAACTGATATCTGTTGTCGTTTATCGAAGGAATCAGGGAATTACCGAAGAGCAGTTGAAACCAATCTTCGAATTGGCGACCAAAATCCGAGGACCCCAACGATTGCCGAAAATCTGCGATAATCCCTTGAAAGATATTTTGCTCCAATAG
Protein
MYRGILVFLTIYLTTSEAVTNSTNCGYFQADFDLISALGTWHVVAIIPEKLFPDKDVTCYKMEISETDEAGLRWLINRTTHGPQNKSIIADVKGTIVRQRYHTEHPFDVWSKSTAGIKGCFQQVLSLDLDKNRIHKTLAHDAMMQLHIVDSKDGSSPFLMQMLWGKLISVVVYRRNQGITEEQLKPIFELATKIRGPQRLPKICDNPLKDILLQ
Summary
Similarity
Belongs to the calycin superfamily. Lipocalin family.
Uniprot
EMBL
Proteomes
PRIDE
SUPFAM
SSF50814
SSF50814
Gene 3D
ProteinModelPortal
Ontologies
GO
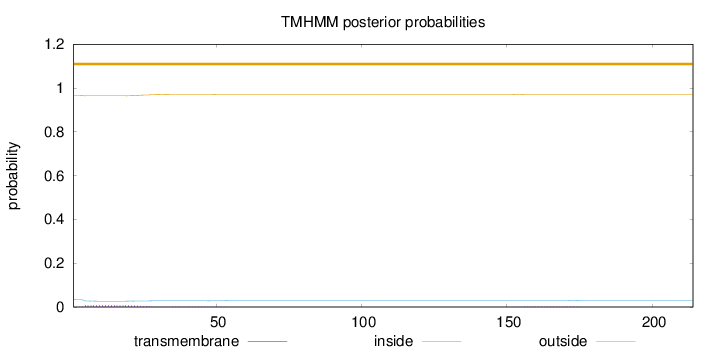
Topology
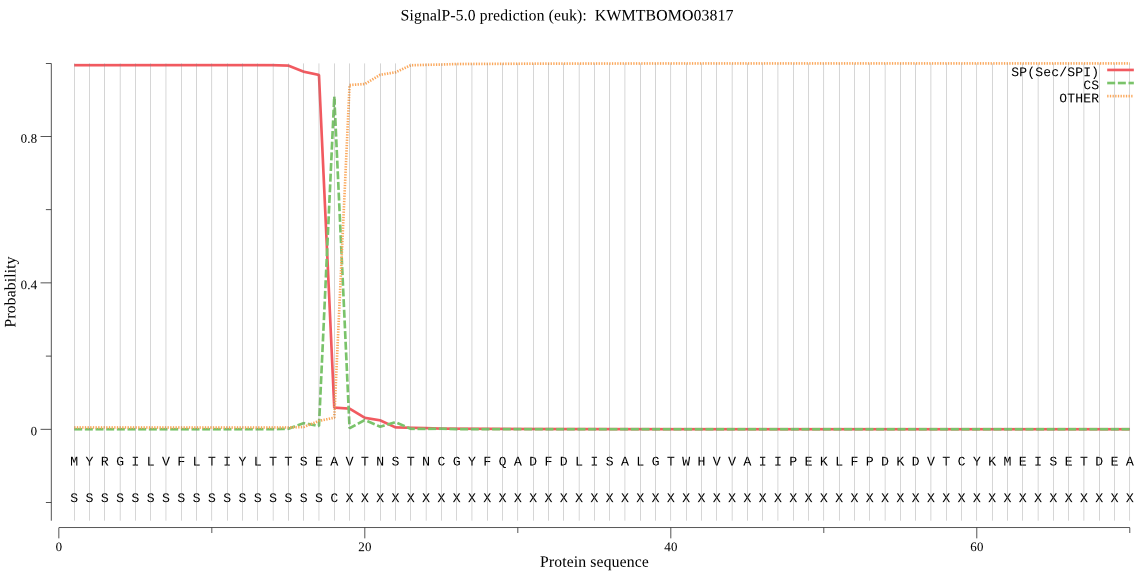
SignalP
Position: 1 - 18,
Likelihood: 0.994939
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24389
Exp number, first 60 AAs:
0.21258
Total prob of N-in:
0.03406
outside
1 - 214
Population Genetic Test Statistics
Pi
346.821659
Theta
238.58014
Tajima's D
1.405544
CLR
0.047388
CSRT
0.768561571921404
Interpretation
Uncertain
