Gene
KWMTBOMO03815
Pre Gene Modal
BGIBMGA010084
Annotation
hypothetical_protein_RR48_07712_[Papilio_machaon]
Full name
MICOS complex subunit MIC10
Location in the cell
Mitochondrial Reliability : 1.07
Sequence
CDS
ATGAGTAAAACCGACAAACGTGACTTTGAAGATCGGTACTCTGCATGTTTCGTTGATTTCGGATTGAAAACAGTGACAGGGCTCTTAATAGGCAGCATGATGGGTAGTTTCTTCTTGAGAGGCTACAAGAAATGGCCAATGTACATCGGCGGCGGTCTCGGTTTCGGGATGGCGTATTCTAATTGCGAGAACAGCCTCAACGACTATTTATTGGCAATGAATCCCAAGCCTTGTTCTATCAAGTTAGTATGA
Protein
MSKTDKRDFEDRYSACFVDFGLKTVTGLLIGSMMGSFFLRGYKKWPMYIGGGLGFGMAYSNCENSLNDYLLAMNPKPCSIKLV
Summary
Description
Component of the MICOS complex, a large protein complex of the mitochondrial inner membrane that plays crucial roles in the maintenance of crista junctions, inner membrane architecture, and formation of contact sites to the outer membrane.
Subunit
Component of the mitochondrial contact site and cristae organizing system (MICOS) complex.
Similarity
Belongs to the MICOS complex subunit Mic10 family.
Uniprot
H9JKN3
A0A3S2P725
A0A194R4C4
A0A0N0PAR7
A0A2P8YGV1
S4NI77
+ More
A0A158NBT6 A0A3S2LJ30 A0A195FD99 K7IZ00 F4X8A2 H2XYB2 A0A088A8Q1 A0A151X9F6 A0A195C9R3 A0A2A3EGQ0 V9IGX3 A0A151J9L9 A0A0J7L4A0 B4R4M1 B4IG99 Q9VY65 A0A310S9D9 A0A0N1IN63 F3YD88 A0A1W4VGA0 A0A3P9K7H3 A0A3Q3T3G2 A0A232FJV3 H9J045 A0A3Q3NHE9 E2C433 J3JZJ1 A0A1I8PJA5 B3MZT7 B4Q2Q0 B3NVW6 A0A3B3H3D2 A0A1I8MJN0 T1HM68 A0A3P9JNH9 A0A1L8EFD6 T1JLT9 A0A3Q2UV46 A0A3P9BY69 A0A3B3DNE3 A0A3B3DLH3 A0A3Q1HDK7 A0A3Q4GG42 A0A3B3Z8A7 A0A3S2PEF4 A0A3Q4MEP0 A0A3Q3IWN7 A0A3B1IHU7 J9LSD1 A0A0L7K3N2 A0A3B5L215 A0A3B3C7R7 A0A3Q1EDK5 A0A3B5QRY2 A0A3Q2NX60 A0A3Q1F2Q7 A0A146P5U2 A0A2I4B223 A0A3Q3FJ21 A0A3Q3RXW6 W5JU03 A0A2U9C013 A0A0N5DLL6 A0A2M3Z6F9 A0A182FJI1 A0A2M4C6E8 A0A336M7C6 T1E7H1 A0A3Q3K1C3 A0A3P8UNP8 A0A3Q3XI66 A8E6W1 E9IJG9 A0A437D5K2 A0A3Q3FGX6 A0A3B4ZGY9 A0A3Q2DJE9 A0A3Q1HPG4
A0A158NBT6 A0A3S2LJ30 A0A195FD99 K7IZ00 F4X8A2 H2XYB2 A0A088A8Q1 A0A151X9F6 A0A195C9R3 A0A2A3EGQ0 V9IGX3 A0A151J9L9 A0A0J7L4A0 B4R4M1 B4IG99 Q9VY65 A0A310S9D9 A0A0N1IN63 F3YD88 A0A1W4VGA0 A0A3P9K7H3 A0A3Q3T3G2 A0A232FJV3 H9J045 A0A3Q3NHE9 E2C433 J3JZJ1 A0A1I8PJA5 B3MZT7 B4Q2Q0 B3NVW6 A0A3B3H3D2 A0A1I8MJN0 T1HM68 A0A3P9JNH9 A0A1L8EFD6 T1JLT9 A0A3Q2UV46 A0A3P9BY69 A0A3B3DNE3 A0A3B3DLH3 A0A3Q1HDK7 A0A3Q4GG42 A0A3B3Z8A7 A0A3S2PEF4 A0A3Q4MEP0 A0A3Q3IWN7 A0A3B1IHU7 J9LSD1 A0A0L7K3N2 A0A3B5L215 A0A3B3C7R7 A0A3Q1EDK5 A0A3B5QRY2 A0A3Q2NX60 A0A3Q1F2Q7 A0A146P5U2 A0A2I4B223 A0A3Q3FJ21 A0A3Q3RXW6 W5JU03 A0A2U9C013 A0A0N5DLL6 A0A2M3Z6F9 A0A182FJI1 A0A2M4C6E8 A0A336M7C6 T1E7H1 A0A3Q3K1C3 A0A3P8UNP8 A0A3Q3XI66 A8E6W1 E9IJG9 A0A437D5K2 A0A3Q3FGX6 A0A3B4ZGY9 A0A3Q2DJE9 A0A3Q1HPG4
Pubmed
19121390
26354079
29403074
23622113
21347285
20075255
+ More
21719571 12481130 15114417 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17554307 28648823 20798317 22516182 23537049 17550304 25315136 25186727 29451363 25463417 25329095 26227816 23542700 20920257 23761445 24487278 21282665
21719571 12481130 15114417 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17554307 28648823 20798317 22516182 23537049 17550304 25315136 25186727 29451363 25463417 25329095 26227816 23542700 20920257 23761445 24487278 21282665
EMBL
BABH01024853
RSAL01000259
RVE43366.1
KQ460930
KPJ10706.1
KQ459167
+ More
KQ458540 KPJ03495.1 KPJ05833.1 PYGN01000604 PSN43490.1 GAIX01014184 JAA78376.1 ADTU01011317 RSAL01000002 RVE54951.1 KQ981673 KYN38378.1 GL888932 EGI57176.1 EAAA01002028 KQ982373 KYQ57025.1 KQ978068 KYM97557.1 KZ288254 PBC30890.1 JR044950 AEY59922.1 KQ979433 KYN21589.1 LBMM01000693 KMQ97732.1 CM000366 EDX17875.1 CH480835 EDW48833.1 AE014298 KX532131 AAF48340.1 ANY27941.1 KQ767320 OAD53408.1 KQ459472 KQ458667 KPJ00175.1 KPJ05591.1 KPJ05592.1 BT126249 AEB33536.1 NNAY01000120 OXU30779.1 BABH01018937 GL452364 EFN77419.1 APGK01059072 BT128673 KB741292 KB632349 AEE63630.1 ENN70396.1 ERL93214.1 CH902635 EDV33888.1 CM000162 EDX01644.1 CH954180 EDV47131.1 ACPB03017303 GFDG01001500 JAV17299.1 JH431581 CM012441 RVE73502.1 ABLF02040834 JTDY01011733 KOB53759.1 GCES01147612 JAQ38710.1 ADMH02000457 ETN66410.1 CP026253 AWP09280.1 GGFM01003340 MBW24091.1 GGFJ01011397 MBW60538.1 UFQT01000496 SSX24769.1 GAMD01003057 JAA98533.1 BT030903 ABV82285.1 GL763802 EFZ19289.1 CM012443 RVE70138.1
KQ458540 KPJ03495.1 KPJ05833.1 PYGN01000604 PSN43490.1 GAIX01014184 JAA78376.1 ADTU01011317 RSAL01000002 RVE54951.1 KQ981673 KYN38378.1 GL888932 EGI57176.1 EAAA01002028 KQ982373 KYQ57025.1 KQ978068 KYM97557.1 KZ288254 PBC30890.1 JR044950 AEY59922.1 KQ979433 KYN21589.1 LBMM01000693 KMQ97732.1 CM000366 EDX17875.1 CH480835 EDW48833.1 AE014298 KX532131 AAF48340.1 ANY27941.1 KQ767320 OAD53408.1 KQ459472 KQ458667 KPJ00175.1 KPJ05591.1 KPJ05592.1 BT126249 AEB33536.1 NNAY01000120 OXU30779.1 BABH01018937 GL452364 EFN77419.1 APGK01059072 BT128673 KB741292 KB632349 AEE63630.1 ENN70396.1 ERL93214.1 CH902635 EDV33888.1 CM000162 EDX01644.1 CH954180 EDV47131.1 ACPB03017303 GFDG01001500 JAV17299.1 JH431581 CM012441 RVE73502.1 ABLF02040834 JTDY01011733 KOB53759.1 GCES01147612 JAQ38710.1 ADMH02000457 ETN66410.1 CP026253 AWP09280.1 GGFM01003340 MBW24091.1 GGFJ01011397 MBW60538.1 UFQT01000496 SSX24769.1 GAMD01003057 JAA98533.1 BT030903 ABV82285.1 GL763802 EFZ19289.1 CM012443 RVE70138.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000245037
UP000005205
+ More
UP000078541 UP000002358 UP000007755 UP000008144 UP000005203 UP000075809 UP000078542 UP000242457 UP000078492 UP000036403 UP000000304 UP000001292 UP000000803 UP000192221 UP000265180 UP000261640 UP000215335 UP000008237 UP000019118 UP000030742 UP000095300 UP000007801 UP000002282 UP000008711 UP000001038 UP000095301 UP000015103 UP000265200 UP000264840 UP000265160 UP000261560 UP000265040 UP000261580 UP000261520 UP000261600 UP000018467 UP000007819 UP000037510 UP000261380 UP000257200 UP000002852 UP000265000 UP000192220 UP000261660 UP000000673 UP000246464 UP000046395 UP000069272 UP000265120 UP000261620 UP000261400 UP000265020
UP000078541 UP000002358 UP000007755 UP000008144 UP000005203 UP000075809 UP000078542 UP000242457 UP000078492 UP000036403 UP000000304 UP000001292 UP000000803 UP000192221 UP000265180 UP000261640 UP000215335 UP000008237 UP000019118 UP000030742 UP000095300 UP000007801 UP000002282 UP000008711 UP000001038 UP000095301 UP000015103 UP000265200 UP000264840 UP000265160 UP000261560 UP000265040 UP000261580 UP000261520 UP000261600 UP000018467 UP000007819 UP000037510 UP000261380 UP000257200 UP000002852 UP000265000 UP000192220 UP000261660 UP000000673 UP000246464 UP000046395 UP000069272 UP000265120 UP000261620 UP000261400 UP000265020
Pfam
PF04418 DUF543
Interpro
IPR007512
Mic10
ProteinModelPortal
H9JKN3
A0A3S2P725
A0A194R4C4
A0A0N0PAR7
A0A2P8YGV1
S4NI77
+ More
A0A158NBT6 A0A3S2LJ30 A0A195FD99 K7IZ00 F4X8A2 H2XYB2 A0A088A8Q1 A0A151X9F6 A0A195C9R3 A0A2A3EGQ0 V9IGX3 A0A151J9L9 A0A0J7L4A0 B4R4M1 B4IG99 Q9VY65 A0A310S9D9 A0A0N1IN63 F3YD88 A0A1W4VGA0 A0A3P9K7H3 A0A3Q3T3G2 A0A232FJV3 H9J045 A0A3Q3NHE9 E2C433 J3JZJ1 A0A1I8PJA5 B3MZT7 B4Q2Q0 B3NVW6 A0A3B3H3D2 A0A1I8MJN0 T1HM68 A0A3P9JNH9 A0A1L8EFD6 T1JLT9 A0A3Q2UV46 A0A3P9BY69 A0A3B3DNE3 A0A3B3DLH3 A0A3Q1HDK7 A0A3Q4GG42 A0A3B3Z8A7 A0A3S2PEF4 A0A3Q4MEP0 A0A3Q3IWN7 A0A3B1IHU7 J9LSD1 A0A0L7K3N2 A0A3B5L215 A0A3B3C7R7 A0A3Q1EDK5 A0A3B5QRY2 A0A3Q2NX60 A0A3Q1F2Q7 A0A146P5U2 A0A2I4B223 A0A3Q3FJ21 A0A3Q3RXW6 W5JU03 A0A2U9C013 A0A0N5DLL6 A0A2M3Z6F9 A0A182FJI1 A0A2M4C6E8 A0A336M7C6 T1E7H1 A0A3Q3K1C3 A0A3P8UNP8 A0A3Q3XI66 A8E6W1 E9IJG9 A0A437D5K2 A0A3Q3FGX6 A0A3B4ZGY9 A0A3Q2DJE9 A0A3Q1HPG4
A0A158NBT6 A0A3S2LJ30 A0A195FD99 K7IZ00 F4X8A2 H2XYB2 A0A088A8Q1 A0A151X9F6 A0A195C9R3 A0A2A3EGQ0 V9IGX3 A0A151J9L9 A0A0J7L4A0 B4R4M1 B4IG99 Q9VY65 A0A310S9D9 A0A0N1IN63 F3YD88 A0A1W4VGA0 A0A3P9K7H3 A0A3Q3T3G2 A0A232FJV3 H9J045 A0A3Q3NHE9 E2C433 J3JZJ1 A0A1I8PJA5 B3MZT7 B4Q2Q0 B3NVW6 A0A3B3H3D2 A0A1I8MJN0 T1HM68 A0A3P9JNH9 A0A1L8EFD6 T1JLT9 A0A3Q2UV46 A0A3P9BY69 A0A3B3DNE3 A0A3B3DLH3 A0A3Q1HDK7 A0A3Q4GG42 A0A3B3Z8A7 A0A3S2PEF4 A0A3Q4MEP0 A0A3Q3IWN7 A0A3B1IHU7 J9LSD1 A0A0L7K3N2 A0A3B5L215 A0A3B3C7R7 A0A3Q1EDK5 A0A3B5QRY2 A0A3Q2NX60 A0A3Q1F2Q7 A0A146P5U2 A0A2I4B223 A0A3Q3FJ21 A0A3Q3RXW6 W5JU03 A0A2U9C013 A0A0N5DLL6 A0A2M3Z6F9 A0A182FJI1 A0A2M4C6E8 A0A336M7C6 T1E7H1 A0A3Q3K1C3 A0A3P8UNP8 A0A3Q3XI66 A8E6W1 E9IJG9 A0A437D5K2 A0A3Q3FGX6 A0A3B4ZGY9 A0A3Q2DJE9 A0A3Q1HPG4
Ontologies
PANTHER
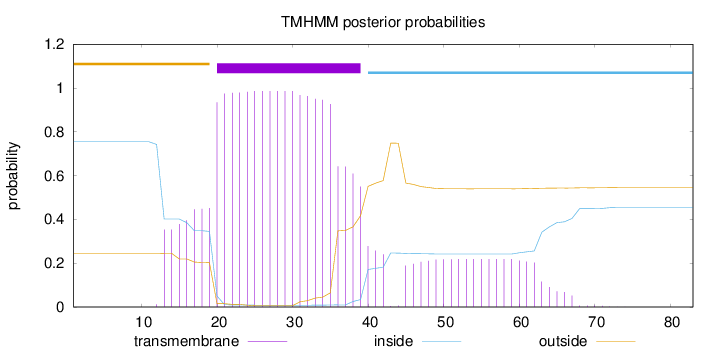
Topology
Subcellular location
Mitochondrion inner membrane
Length:
83
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.83774
Exp number, first 60 AAs:
25.00305
Total prob of N-in:
0.75450
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 39
inside
40 - 83
Population Genetic Test Statistics
Pi
263.92161
Theta
174.094107
Tajima's D
1.503096
CLR
0
CSRT
0.790960451977401
Interpretation
Uncertain
