Gene
KWMTBOMO03808
Pre Gene Modal
BGIBMGA010081
Annotation
PREDICTED:_protein_tipE_isoform_X1_[Bombyx_mori]
Full name
Protein tipE
Alternative Name
Temperature-induced paralytic E
Location in the cell
PlasmaMembrane Reliability : 3.072
Sequence
CDS
ATGGCCGACGAAGAGAAAATCCCTCCGACCTTTCTTGAGAAACTCCTATTCTACACAACAGCCTCCTTCGTTCTTCTTGCCACGTTCAGCCTATTCGCCTTTCTATTCTTAGTGCCATTTGTGATTGAACCAGCTTTCACTACGATATTCATGCAATTCGATCCTATAGGAGCTTTGTGTGTTACCACCCAAGTTAAACATTTGGTCGGAGCGAGCAATTGCACGTGGGCGTCCTGCAGGGAGGGCTGCACTAAAGATCTATTTGAATGCACACAAATTAGGGTCAGCTATAAACTGGGTAATTCATCTAATGTTACTGAAGAGGAAGTTGAAAATCTTATCAGAGTAGAGAGGGCTCTCAGGAATGATTACGACTATGAGAACGAGGTAGCTGCTGATAAGTCTTATCCAGATATGGGCGAGAGGGATGACTTGCCTGAACCGTATCCAACCGGGCTACAAGGGAACGACTCGGAATGGTATTTTACCGGGGCCCGTTTGTTTCCTAACGTGAAAGGTTGCGGTTATCCTCCTATATTGAACTGCACAATATTCTACGGAAAATATAGGCCTATTGGCACCAACTTCACTTGCTACTATAGCCGGGTGGATCCGGGCTTGGTTATCACCGAACTGGACATGTGGCAGAACACTCTGAACTTGGTGTACGCGATGGCCATACCAATACCGTCATTCTTTATCTCCGTCATATATTTGACGTTCGCTTACTTCAAAATTTATAATACTGAAGATGAATCCGAGCAGGCGCCTCTGAAGAGTGACGCTGAGGCTATGGCCACCGGTGACGATGAGAAACCAACGACCCCAGGGTCTGATACGTTTAGAGAAGATCTCGCATGTTTCGGTCACCAATTAAAGATGCAACTAGCCAACGATAAGCCTAAAGACGGCTTCGAGTCCAATAGTCCACCCATTTCCAATTCTGCCTCACTGTCTGGGACCAAGTCGTCATTCGTGAACGCCTAA
Protein
MADEEKIPPTFLEKLLFYTTASFVLLATFSLFAFLFLVPFVIEPAFTTIFMQFDPIGALCVTTQVKHLVGASNCTWASCREGCTKDLFECTQIRVSYKLGNSSNVTEEEVENLIRVERALRNDYDYENEVAADKSYPDMGERDDLPEPYPTGLQGNDSEWYFTGARLFPNVKGCGYPPILNCTIFYGKYRPIGTNFTCYYSRVDPGLVITELDMWQNTLNLVYAMAIPIPSFFISVIYLTFAYFKIYNTEDESEQAPLKSDAEAMATGDDEKPTTPGSDTFREDLACFGHQLKMQLANDKPKDGFESNSPPISNSASLSGTKSSFVNA
Summary
Description
Enhances para sodium channel function. Required during pupal development to rescue adult paralysis and also protects adult flies against heat-induced lethality.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Protein tipE
Uniprot
H9JKN0
A0A3S2LUB4
A0A2H1VE86
A0A2A4JVK2
A0A1C9HJF5
A0A212F9B3
+ More
A0A194QEW5 A0A194RNY5 A0A0L7L9Y4 A0A2A4JUK1 U3GPB8 A0A1W4WKP3 A0A1B6DPF9 A0A2J7RPJ4 A0A310SHL1 A0A067QXZ1 A0A0Y0GW99 A0A088A7P9 A0A1Y1LN36 A0A3L8DZA7 E2BUU4 A0A158NJJ8 E9I9B6 A0A2P8XF56 E2A570 A0A2A3ENP1 A0A195F1J4 A0A151WJ86 A0A154PAU7 N6U6I2 A0A0A9XCZ5 A0A0A9XHV8 D6W8C5 T1HDH6 A0A336LZL0 A0A1B0CX46 A0A232F3B8 V9ID25 B4LB99 A0A1J1HWK3 E0VWJ9 P48613 A0A1W4W659 A0A195BFR0 B4HU17 A0A0J9RNL4 B3NG52 B4PHR2 B3M4W6 A0A1S3D2V6 A0A3B0K235 B4H7W4 Q29D71 A0A195CAH4 B4L9C6 B4J1F7 A0A1I8PYK2 A0A1I8PYM6 B4QQH2 A0A2S2NYM8 X1WP79 A0A1V0CJZ5 A0A0P5YZD7
A0A194QEW5 A0A194RNY5 A0A0L7L9Y4 A0A2A4JUK1 U3GPB8 A0A1W4WKP3 A0A1B6DPF9 A0A2J7RPJ4 A0A310SHL1 A0A067QXZ1 A0A0Y0GW99 A0A088A7P9 A0A1Y1LN36 A0A3L8DZA7 E2BUU4 A0A158NJJ8 E9I9B6 A0A2P8XF56 E2A570 A0A2A3ENP1 A0A195F1J4 A0A151WJ86 A0A154PAU7 N6U6I2 A0A0A9XCZ5 A0A0A9XHV8 D6W8C5 T1HDH6 A0A336LZL0 A0A1B0CX46 A0A232F3B8 V9ID25 B4LB99 A0A1J1HWK3 E0VWJ9 P48613 A0A1W4W659 A0A195BFR0 B4HU17 A0A0J9RNL4 B3NG52 B4PHR2 B3M4W6 A0A1S3D2V6 A0A3B0K235 B4H7W4 Q29D71 A0A195CAH4 B4L9C6 B4J1F7 A0A1I8PYK2 A0A1I8PYM6 B4QQH2 A0A2S2NYM8 X1WP79 A0A1V0CJZ5 A0A0P5YZD7
Pubmed
EMBL
BABH01024828
RSAL01000215
RVE44136.1
ODYU01002075
SOQ39127.1
NWSH01000549
+ More
PCG75718.1 KU739069 AOO86803.1 AGBW02009597 OWR50345.1 KQ459167 KPJ03490.1 KQ460118 KPJ17731.1 JTDY01002018 KOB72313.1 PCG75717.1 KC992733 AGT98549.1 GEDC01009751 JAS27547.1 NEVH01001358 PNF42753.1 KQ761164 OAD58097.1 KK852829 KDR15345.1 KT455381 AMB38676.1 GEZM01054903 JAV73345.1 QOIP01000003 RLU25138.1 GL450757 EFN80530.1 ADTU01017981 GL761765 EFZ22843.1 PYGN01002378 PSN30612.1 GL436875 EFN71413.1 KZ288203 PBC33337.1 KQ981856 KYN34445.1 KQ983045 KYQ47891.1 KQ434849 KZC08504.1 APGK01047144 KB741077 KB632384 ENN74162.1 ERL94296.1 GBHO01026951 JAG16653.1 GBHO01026949 GBRD01002376 GDHC01019041 JAG16655.1 JAG63445.1 JAP99587.1 KQ971307 EFA10931.2 ACPB03015640 UFQS01000257 UFQT01000257 SSX02084.1 SSX22461.1 AJWK01033191 AJWK01033192 NNAY01001069 OXU25254.1 JR038322 AEY58209.1 CH940647 EDW68663.2 KRF83962.1 KRF83963.1 KRF83964.1 CVRI01000029 CRK92487.1 DS235822 EEB17755.1 U27561 AE014296 AY060317 KQ976490 KYM83420.1 CH480817 EDW50438.1 CM002912 KMY97525.1 KMY97526.1 KMY97527.1 KMY97528.1 KMY97529.1 KMY97530.1 CH954178 EDV50881.1 KQS43564.1 KQS43565.1 KQS43566.1 KQS43567.1 CM000159 EDW93371.1 KRK01140.1 KRK01141.1 KRK01142.1 KRK01143.1 KRK01144.1 CH902618 EDV40540.1 KPU78623.1 OUUW01000002 SPP77448.1 CH479219 EDW34754.1 CH379070 EAL30543.1 KRT09039.1 KRT09040.1 KQ978081 KYM97113.1 CH934399 CH933816 EDW05404.1 EDW17301.1 KRG07718.1 CH916366 EDV95848.1 CM000363 EDX09178.1 GGMR01009668 MBY22287.1 ABLF02030064 KY476317 ARA68389.1 GDIP01051261 JAM52454.1
PCG75718.1 KU739069 AOO86803.1 AGBW02009597 OWR50345.1 KQ459167 KPJ03490.1 KQ460118 KPJ17731.1 JTDY01002018 KOB72313.1 PCG75717.1 KC992733 AGT98549.1 GEDC01009751 JAS27547.1 NEVH01001358 PNF42753.1 KQ761164 OAD58097.1 KK852829 KDR15345.1 KT455381 AMB38676.1 GEZM01054903 JAV73345.1 QOIP01000003 RLU25138.1 GL450757 EFN80530.1 ADTU01017981 GL761765 EFZ22843.1 PYGN01002378 PSN30612.1 GL436875 EFN71413.1 KZ288203 PBC33337.1 KQ981856 KYN34445.1 KQ983045 KYQ47891.1 KQ434849 KZC08504.1 APGK01047144 KB741077 KB632384 ENN74162.1 ERL94296.1 GBHO01026951 JAG16653.1 GBHO01026949 GBRD01002376 GDHC01019041 JAG16655.1 JAG63445.1 JAP99587.1 KQ971307 EFA10931.2 ACPB03015640 UFQS01000257 UFQT01000257 SSX02084.1 SSX22461.1 AJWK01033191 AJWK01033192 NNAY01001069 OXU25254.1 JR038322 AEY58209.1 CH940647 EDW68663.2 KRF83962.1 KRF83963.1 KRF83964.1 CVRI01000029 CRK92487.1 DS235822 EEB17755.1 U27561 AE014296 AY060317 KQ976490 KYM83420.1 CH480817 EDW50438.1 CM002912 KMY97525.1 KMY97526.1 KMY97527.1 KMY97528.1 KMY97529.1 KMY97530.1 CH954178 EDV50881.1 KQS43564.1 KQS43565.1 KQS43566.1 KQS43567.1 CM000159 EDW93371.1 KRK01140.1 KRK01141.1 KRK01142.1 KRK01143.1 KRK01144.1 CH902618 EDV40540.1 KPU78623.1 OUUW01000002 SPP77448.1 CH479219 EDW34754.1 CH379070 EAL30543.1 KRT09039.1 KRT09040.1 KQ978081 KYM97113.1 CH934399 CH933816 EDW05404.1 EDW17301.1 KRG07718.1 CH916366 EDV95848.1 CM000363 EDX09178.1 GGMR01009668 MBY22287.1 ABLF02030064 KY476317 ARA68389.1 GDIP01051261 JAM52454.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000037510 UP000192223 UP000235965 UP000027135 UP000005203 UP000279307 UP000008237 UP000005205 UP000245037 UP000000311 UP000242457 UP000078541 UP000075809 UP000076502 UP000019118 UP000030742 UP000007266 UP000015103 UP000092461 UP000215335 UP000008792 UP000183832 UP000009046 UP000000803 UP000192221 UP000078540 UP000001292 UP000008711 UP000002282 UP000007801 UP000079169 UP000268350 UP000008744 UP000001819 UP000078542 UP000009192 UP000001070 UP000095300 UP000000304 UP000007819
UP000037510 UP000192223 UP000235965 UP000027135 UP000005203 UP000279307 UP000008237 UP000005205 UP000245037 UP000000311 UP000242457 UP000078541 UP000075809 UP000076502 UP000019118 UP000030742 UP000007266 UP000015103 UP000092461 UP000215335 UP000008792 UP000183832 UP000009046 UP000000803 UP000192221 UP000078540 UP000001292 UP000008711 UP000002282 UP000007801 UP000079169 UP000268350 UP000008744 UP000001819 UP000078542 UP000009192 UP000001070 UP000095300 UP000000304 UP000007819
ProteinModelPortal
H9JKN0
A0A3S2LUB4
A0A2H1VE86
A0A2A4JVK2
A0A1C9HJF5
A0A212F9B3
+ More
A0A194QEW5 A0A194RNY5 A0A0L7L9Y4 A0A2A4JUK1 U3GPB8 A0A1W4WKP3 A0A1B6DPF9 A0A2J7RPJ4 A0A310SHL1 A0A067QXZ1 A0A0Y0GW99 A0A088A7P9 A0A1Y1LN36 A0A3L8DZA7 E2BUU4 A0A158NJJ8 E9I9B6 A0A2P8XF56 E2A570 A0A2A3ENP1 A0A195F1J4 A0A151WJ86 A0A154PAU7 N6U6I2 A0A0A9XCZ5 A0A0A9XHV8 D6W8C5 T1HDH6 A0A336LZL0 A0A1B0CX46 A0A232F3B8 V9ID25 B4LB99 A0A1J1HWK3 E0VWJ9 P48613 A0A1W4W659 A0A195BFR0 B4HU17 A0A0J9RNL4 B3NG52 B4PHR2 B3M4W6 A0A1S3D2V6 A0A3B0K235 B4H7W4 Q29D71 A0A195CAH4 B4L9C6 B4J1F7 A0A1I8PYK2 A0A1I8PYM6 B4QQH2 A0A2S2NYM8 X1WP79 A0A1V0CJZ5 A0A0P5YZD7
A0A194QEW5 A0A194RNY5 A0A0L7L9Y4 A0A2A4JUK1 U3GPB8 A0A1W4WKP3 A0A1B6DPF9 A0A2J7RPJ4 A0A310SHL1 A0A067QXZ1 A0A0Y0GW99 A0A088A7P9 A0A1Y1LN36 A0A3L8DZA7 E2BUU4 A0A158NJJ8 E9I9B6 A0A2P8XF56 E2A570 A0A2A3ENP1 A0A195F1J4 A0A151WJ86 A0A154PAU7 N6U6I2 A0A0A9XCZ5 A0A0A9XHV8 D6W8C5 T1HDH6 A0A336LZL0 A0A1B0CX46 A0A232F3B8 V9ID25 B4LB99 A0A1J1HWK3 E0VWJ9 P48613 A0A1W4W659 A0A195BFR0 B4HU17 A0A0J9RNL4 B3NG52 B4PHR2 B3M4W6 A0A1S3D2V6 A0A3B0K235 B4H7W4 Q29D71 A0A195CAH4 B4L9C6 B4J1F7 A0A1I8PYK2 A0A1I8PYM6 B4QQH2 A0A2S2NYM8 X1WP79 A0A1V0CJZ5 A0A0P5YZD7
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
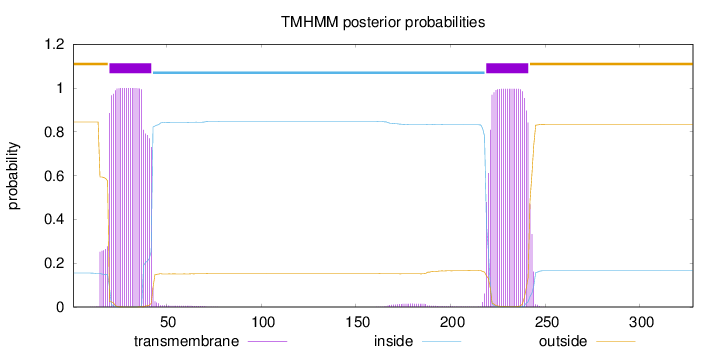
Length:
328
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
46.18627
Exp number, first 60 AAs:
23.23509
Total prob of N-in:
0.15481
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 218
TMhelix
219 - 241
outside
242 - 328
Population Genetic Test Statistics
Pi
288.582645
Theta
207.680042
Tajima's D
1.36796
CLR
0.364004
CSRT
0.758562071896405
Interpretation
Uncertain
