Gene
KWMTBOMO03807
Pre Gene Modal
BGIBMGA010080
Annotation
PREDICTED:_protein_tipE_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.958
Sequence
CDS
ATGGGCGGTTCAGCAGCGCCCGGTAGTCCGGAATTTAAGCCCCTGGAACCGACAAGAGAGCAATTATTAGCCCACTTCTTTGAGAGATTCAAATTTTACACGTCACTTTGCCTCGGCACCGCGGCCATATTGGCCGTTTTCGCTTTTCTATTCCTGATACCGTTCGTAGTTGAGCCGGCCGTGCAGACGATATTAGCTGAGTTCGTGCCGGAAGCCGGTCTATGCTCTGTGTCCGAGCACGTTCACGCCGAGACGCTGACTAATTGCACGTGGGCGTCGTGTAGAGAAGGCTGCACAACCGCACACACAAAGTGTCACAAGATACGAGTGAACTTGTCGCGTAGGCCGTTTGTCGAGGGAGCGCCGCCCCCAATCAAATGGGACGAGACGCACTTGAAGTTCCTAATCAACCCTGAGGGATGCGGCTACCCTCCAACTGTCAACTGCTCGGTGTTTGCGAAAGAATACGCCGGCCCAAACGCTCCAAAGCTGTTCCCTTGTTACTACAGTTTGACTAGGCCAGATCTCGTGGTCGCGAGGTATTCGTGGGAGGAGACCGTGCGAGGGCTGATCATTGCTCTGGTGCTACCGACCTCCGTGTTCGTGTTCAGTTTAAGTGTACTCGCATATTGGCACTGTCGATGTTGCGACCGGGCCTGCAATCGACGAGTTCACGCCGAATCATTTGCGAGCAAGGAAGATAGCAAGTTACTGTGTGAGTTAGATGACGATCCAAGTGATGACGTGTTCTGA
Protein
MGGSAAPGSPEFKPLEPTREQLLAHFFERFKFYTSLCLGTAAILAVFAFLFLIPFVVEPAVQTILAEFVPEAGLCSVSEHVHAETLTNCTWASCREGCTTAHTKCHKIRVNLSRRPFVEGAPPPIKWDETHLKFLINPEGCGYPPTVNCSVFAKEYAGPNAPKLFPCYYSLTRPDLVVARYSWEETVRGLIIALVLPTSVFVFSLSVLAYWHCRCCDRACNRRVHAESFASKEDSKLLCELDDDPSDDVF
Summary
Uniprot
H9JKM9
A0A3S2N895
A0A2A4JVB5
A0A2H1V514
A0A212EXA3
A0A194RJK2
+ More
A0A194QDJ3 A0A1C9HJF0 A0A0L7L909 Q17J69 A0A182G4I6 A0A182XHQ3 Q7QCV4 A0A182TWK8 A0A0K8V914 A0A034W2H3 A0A182W004 A0A182QWN7 A0A182MNC8 A0A1Q3FVU9 W5JGB2 B4L9C9 B3M4W3 A0A1W4W5C3 B4PHQ9 B4HU14 B3NG49 B4QQG9 B4J1F4 B4N3H0 A0A182IL37 Q9VZG9 B4LB95 Q29D74 A0A3B0J6T3 A0A023ENS8 A0A182G526 A0A1J1IL75 A0A182VH04 A0A182Y450 A0A182SX04 A0A182RNX2 A0A182P269 A0A182HTN5 A0A1I8PAP5 A0A195C8T2 A0A1Q3FLT9 A0A1A9ZKD2 A0A1A9UG48 W8BJR4 A0A1I8N706 A0A1A9YIH2 A0A1B0AQU8 A0A0P8XV64 W8AYC6 A0A195BGZ1 A0A158NJK0 A0A1W4W6Q4 A0A182FQT2 A0A0J9RP87 A0A0R1DYY2 A0A0Q5U3Y3 A0A195EKJ7 A0A0Q9XTR3 F4X3Y2 A0A0R3P7D5 A0A0Q9WHE5 A0A182LGI0 B6IDT8 A0A0M4EHG4 A0A0P9ALA5 B3DNE8 A0A0R3P7B9 A0A2A3ENJ6 V9IE85 A0A0R3P931 A0A088A836 A0A0R3P767 A0A120HXK1 A0A1B0FH52 E2BUU6 A0A154P990 A0A0L7QRP3 A0A1I8PAW4 A0A026WRF2 E9I9B8 A0A067R9U4 A0A1I8N708 A0A0C9QE29 B0WB57 A0A2P8XF25 E0VWK1 E2A572 A0A0J7P160 A0A2J7RPJ7 A0A2P8XAE9 A0A1W4WJH1 D6W8C3 A0A1Y1K4N7
A0A194QDJ3 A0A1C9HJF0 A0A0L7L909 Q17J69 A0A182G4I6 A0A182XHQ3 Q7QCV4 A0A182TWK8 A0A0K8V914 A0A034W2H3 A0A182W004 A0A182QWN7 A0A182MNC8 A0A1Q3FVU9 W5JGB2 B4L9C9 B3M4W3 A0A1W4W5C3 B4PHQ9 B4HU14 B3NG49 B4QQG9 B4J1F4 B4N3H0 A0A182IL37 Q9VZG9 B4LB95 Q29D74 A0A3B0J6T3 A0A023ENS8 A0A182G526 A0A1J1IL75 A0A182VH04 A0A182Y450 A0A182SX04 A0A182RNX2 A0A182P269 A0A182HTN5 A0A1I8PAP5 A0A195C8T2 A0A1Q3FLT9 A0A1A9ZKD2 A0A1A9UG48 W8BJR4 A0A1I8N706 A0A1A9YIH2 A0A1B0AQU8 A0A0P8XV64 W8AYC6 A0A195BGZ1 A0A158NJK0 A0A1W4W6Q4 A0A182FQT2 A0A0J9RP87 A0A0R1DYY2 A0A0Q5U3Y3 A0A195EKJ7 A0A0Q9XTR3 F4X3Y2 A0A0R3P7D5 A0A0Q9WHE5 A0A182LGI0 B6IDT8 A0A0M4EHG4 A0A0P9ALA5 B3DNE8 A0A0R3P7B9 A0A2A3ENJ6 V9IE85 A0A0R3P931 A0A088A836 A0A0R3P767 A0A120HXK1 A0A1B0FH52 E2BUU6 A0A154P990 A0A0L7QRP3 A0A1I8PAW4 A0A026WRF2 E9I9B8 A0A067R9U4 A0A1I8N708 A0A0C9QE29 B0WB57 A0A2P8XF25 E0VWK1 E2A572 A0A0J7P160 A0A2J7RPJ7 A0A2P8XAE9 A0A1W4WJH1 D6W8C3 A0A1Y1K4N7
Pubmed
19121390
22118469
26354079
26227816
17510324
26483478
+ More
12364791 14747013 17210077 25348373 20920257 23761445 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 16325765 17569856 17569867 26109357 26109356 15632085 24945155 25244985 24495485 25315136 21347285 21719571 20966253 26202396 20798317 24508170 30249741 21282665 24845553 29403074 20566863 18362917 19820115 28004739
12364791 14747013 17210077 25348373 20920257 23761445 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 16325765 17569856 17569867 26109357 26109356 15632085 24945155 25244985 24495485 25315136 21347285 21719571 20966253 26202396 20798317 24508170 30249741 21282665 24845553 29403074 20566863 18362917 19820115 28004739
EMBL
BABH01024824
RSAL01000215
RVE44135.1
NWSH01000549
PCG75716.1
ODYU01000542
+ More
SOQ35472.1 AGBW02011795 OWR46119.1 KQ460118 KPJ17732.1 KQ459167 KPJ03489.1 KU739067 AOO86801.1 JTDY01002239 KOB71824.1 CH477234 EAT46738.1 JXUM01042511 KQ561327 KXJ78971.1 AAAB01008859 EAA07853.5 GDHF01016870 JAI35444.1 GAKP01009196 JAC49756.1 AXCN02000328 AXCM01001634 GFDL01003502 JAV31543.1 ADMH02001605 ETN61859.1 CH933816 EDW17304.1 CH902618 EDV40537.1 CM000159 EDW93368.1 CH480817 EDW50435.1 CH954178 EDV50878.1 CM000363 CM002912 EDX09175.1 KMY97521.1 CH916366 EDV95845.1 CH964095 EDW79175.1 AE014296 DQ139958 AAF47854.1 ABA06529.1 CH940647 EDW68659.1 CH379070 EAL30541.1 OUUW01000002 SPP77445.1 GAPW01003008 JAC10590.1 JXUM01042862 KQ561341 KXJ78915.1 CVRI01000055 CRL00912.1 APCN01000570 KQ978081 KYM97110.1 GFDL01006540 JAV28505.1 GAMC01016696 JAB89859.1 JXJN01002039 JXJN01002040 JXJN01002041 KPU78621.1 GAMC01016697 JAB89858.1 KQ976490 KYM83417.1 ADTU01017981 KMY97522.1 KRK01139.1 KQS43563.1 KQ978782 KYN28442.1 KRG07719.1 GL888624 EGI58929.1 KRT09036.1 KRF83960.1 BT050528 ACJ13235.1 ACZ94617.1 CP012525 ALC44027.1 KPU78622.1 BT032936 ACD99500.1 KRT09035.1 KZ288203 PBC33335.1 JR040105 AEY58977.1 KRT09037.1 KRT09034.1 KT455383 AMB38678.1 CCAG010010924 GL450757 EFN80532.1 KQ434849 KZC08506.1 KQ414782 KOC61171.1 KK107139 QOIP01000003 EZA57669.1 RLU24435.1 GL761765 EFZ22834.1 KK852829 KDR15343.1 GBYB01012663 JAG82430.1 DS231877 EDS42122.1 PYGN01002378 PSN30613.1 DS235822 EEB17757.1 GL436875 EFN71415.1 LBMM01000424 KMQ98390.1 NEVH01001358 PNF42756.1 PYGN01018392 PSN28974.1 KQ971307 EFA10883.1 GEZM01093101 JAV56343.1
SOQ35472.1 AGBW02011795 OWR46119.1 KQ460118 KPJ17732.1 KQ459167 KPJ03489.1 KU739067 AOO86801.1 JTDY01002239 KOB71824.1 CH477234 EAT46738.1 JXUM01042511 KQ561327 KXJ78971.1 AAAB01008859 EAA07853.5 GDHF01016870 JAI35444.1 GAKP01009196 JAC49756.1 AXCN02000328 AXCM01001634 GFDL01003502 JAV31543.1 ADMH02001605 ETN61859.1 CH933816 EDW17304.1 CH902618 EDV40537.1 CM000159 EDW93368.1 CH480817 EDW50435.1 CH954178 EDV50878.1 CM000363 CM002912 EDX09175.1 KMY97521.1 CH916366 EDV95845.1 CH964095 EDW79175.1 AE014296 DQ139958 AAF47854.1 ABA06529.1 CH940647 EDW68659.1 CH379070 EAL30541.1 OUUW01000002 SPP77445.1 GAPW01003008 JAC10590.1 JXUM01042862 KQ561341 KXJ78915.1 CVRI01000055 CRL00912.1 APCN01000570 KQ978081 KYM97110.1 GFDL01006540 JAV28505.1 GAMC01016696 JAB89859.1 JXJN01002039 JXJN01002040 JXJN01002041 KPU78621.1 GAMC01016697 JAB89858.1 KQ976490 KYM83417.1 ADTU01017981 KMY97522.1 KRK01139.1 KQS43563.1 KQ978782 KYN28442.1 KRG07719.1 GL888624 EGI58929.1 KRT09036.1 KRF83960.1 BT050528 ACJ13235.1 ACZ94617.1 CP012525 ALC44027.1 KPU78622.1 BT032936 ACD99500.1 KRT09035.1 KZ288203 PBC33335.1 JR040105 AEY58977.1 KRT09037.1 KRT09034.1 KT455383 AMB38678.1 CCAG010010924 GL450757 EFN80532.1 KQ434849 KZC08506.1 KQ414782 KOC61171.1 KK107139 QOIP01000003 EZA57669.1 RLU24435.1 GL761765 EFZ22834.1 KK852829 KDR15343.1 GBYB01012663 JAG82430.1 DS231877 EDS42122.1 PYGN01002378 PSN30613.1 DS235822 EEB17757.1 GL436875 EFN71415.1 LBMM01000424 KMQ98390.1 NEVH01001358 PNF42756.1 PYGN01018392 PSN28974.1 KQ971307 EFA10883.1 GEZM01093101 JAV56343.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000008820 UP000069940 UP000249989 UP000076407 UP000007062 UP000075902 UP000075920 UP000075886 UP000075883 UP000000673 UP000009192 UP000007801 UP000192221 UP000002282 UP000001292 UP000008711 UP000000304 UP000001070 UP000007798 UP000075880 UP000000803 UP000008792 UP000001819 UP000268350 UP000183832 UP000075903 UP000076408 UP000075901 UP000075900 UP000075885 UP000075840 UP000095300 UP000078542 UP000092445 UP000078200 UP000095301 UP000092443 UP000092460 UP000078540 UP000005205 UP000069272 UP000078492 UP000007755 UP000075882 UP000092553 UP000242457 UP000005203 UP000092444 UP000008237 UP000076502 UP000053825 UP000053097 UP000279307 UP000027135 UP000002320 UP000245037 UP000009046 UP000000311 UP000036403 UP000235965 UP000192223 UP000007266
UP000037510 UP000008820 UP000069940 UP000249989 UP000076407 UP000007062 UP000075902 UP000075920 UP000075886 UP000075883 UP000000673 UP000009192 UP000007801 UP000192221 UP000002282 UP000001292 UP000008711 UP000000304 UP000001070 UP000007798 UP000075880 UP000000803 UP000008792 UP000001819 UP000268350 UP000183832 UP000075903 UP000076408 UP000075901 UP000075900 UP000075885 UP000075840 UP000095300 UP000078542 UP000092445 UP000078200 UP000095301 UP000092443 UP000092460 UP000078540 UP000005205 UP000069272 UP000078492 UP000007755 UP000075882 UP000092553 UP000242457 UP000005203 UP000092444 UP000008237 UP000076502 UP000053825 UP000053097 UP000279307 UP000027135 UP000002320 UP000245037 UP000009046 UP000000311 UP000036403 UP000235965 UP000192223 UP000007266
SUPFAM
SSF50494
SSF50494
ProteinModelPortal
H9JKM9
A0A3S2N895
A0A2A4JVB5
A0A2H1V514
A0A212EXA3
A0A194RJK2
+ More
A0A194QDJ3 A0A1C9HJF0 A0A0L7L909 Q17J69 A0A182G4I6 A0A182XHQ3 Q7QCV4 A0A182TWK8 A0A0K8V914 A0A034W2H3 A0A182W004 A0A182QWN7 A0A182MNC8 A0A1Q3FVU9 W5JGB2 B4L9C9 B3M4W3 A0A1W4W5C3 B4PHQ9 B4HU14 B3NG49 B4QQG9 B4J1F4 B4N3H0 A0A182IL37 Q9VZG9 B4LB95 Q29D74 A0A3B0J6T3 A0A023ENS8 A0A182G526 A0A1J1IL75 A0A182VH04 A0A182Y450 A0A182SX04 A0A182RNX2 A0A182P269 A0A182HTN5 A0A1I8PAP5 A0A195C8T2 A0A1Q3FLT9 A0A1A9ZKD2 A0A1A9UG48 W8BJR4 A0A1I8N706 A0A1A9YIH2 A0A1B0AQU8 A0A0P8XV64 W8AYC6 A0A195BGZ1 A0A158NJK0 A0A1W4W6Q4 A0A182FQT2 A0A0J9RP87 A0A0R1DYY2 A0A0Q5U3Y3 A0A195EKJ7 A0A0Q9XTR3 F4X3Y2 A0A0R3P7D5 A0A0Q9WHE5 A0A182LGI0 B6IDT8 A0A0M4EHG4 A0A0P9ALA5 B3DNE8 A0A0R3P7B9 A0A2A3ENJ6 V9IE85 A0A0R3P931 A0A088A836 A0A0R3P767 A0A120HXK1 A0A1B0FH52 E2BUU6 A0A154P990 A0A0L7QRP3 A0A1I8PAW4 A0A026WRF2 E9I9B8 A0A067R9U4 A0A1I8N708 A0A0C9QE29 B0WB57 A0A2P8XF25 E0VWK1 E2A572 A0A0J7P160 A0A2J7RPJ7 A0A2P8XAE9 A0A1W4WJH1 D6W8C3 A0A1Y1K4N7
A0A194QDJ3 A0A1C9HJF0 A0A0L7L909 Q17J69 A0A182G4I6 A0A182XHQ3 Q7QCV4 A0A182TWK8 A0A0K8V914 A0A034W2H3 A0A182W004 A0A182QWN7 A0A182MNC8 A0A1Q3FVU9 W5JGB2 B4L9C9 B3M4W3 A0A1W4W5C3 B4PHQ9 B4HU14 B3NG49 B4QQG9 B4J1F4 B4N3H0 A0A182IL37 Q9VZG9 B4LB95 Q29D74 A0A3B0J6T3 A0A023ENS8 A0A182G526 A0A1J1IL75 A0A182VH04 A0A182Y450 A0A182SX04 A0A182RNX2 A0A182P269 A0A182HTN5 A0A1I8PAP5 A0A195C8T2 A0A1Q3FLT9 A0A1A9ZKD2 A0A1A9UG48 W8BJR4 A0A1I8N706 A0A1A9YIH2 A0A1B0AQU8 A0A0P8XV64 W8AYC6 A0A195BGZ1 A0A158NJK0 A0A1W4W6Q4 A0A182FQT2 A0A0J9RP87 A0A0R1DYY2 A0A0Q5U3Y3 A0A195EKJ7 A0A0Q9XTR3 F4X3Y2 A0A0R3P7D5 A0A0Q9WHE5 A0A182LGI0 B6IDT8 A0A0M4EHG4 A0A0P9ALA5 B3DNE8 A0A0R3P7B9 A0A2A3ENJ6 V9IE85 A0A0R3P931 A0A088A836 A0A0R3P767 A0A120HXK1 A0A1B0FH52 E2BUU6 A0A154P990 A0A0L7QRP3 A0A1I8PAW4 A0A026WRF2 E9I9B8 A0A067R9U4 A0A1I8N708 A0A0C9QE29 B0WB57 A0A2P8XF25 E0VWK1 E2A572 A0A0J7P160 A0A2J7RPJ7 A0A2P8XAE9 A0A1W4WJH1 D6W8C3 A0A1Y1K4N7
Ontologies
PANTHER
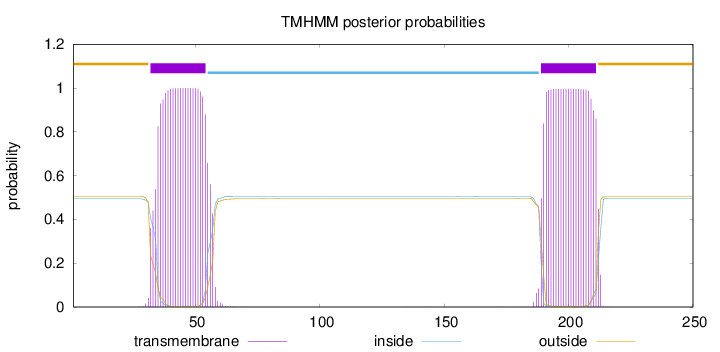
Topology
Length:
250
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.40461
Exp number, first 60 AAs:
22.65374
Total prob of N-in:
0.49427
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 188
TMhelix
189 - 211
outside
212 - 250
Population Genetic Test Statistics
Pi
235.165774
Theta
202.444896
Tajima's D
0.709287
CLR
2.273571
CSRT
0.573171341432928
Interpretation
Uncertain
