Gene
KWMTBOMO03797
Pre Gene Modal
BGIBMGA014330
Annotation
endonuclease-reverse_transcriptase_HmRTE-e01_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.433 Mitochondrial Reliability : 1.108 Nuclear Reliability : 1.596
Sequence
CDS
ATGAGAAGGAATAAAAAGAAGAGCACCAGGGCTAGGGCGTCCCCCGCAGGCGACGCGCAGGGACAGCACCCGGTCTCTGTGGAAAGTGGACGAGGGTTGTCGCATCACGGGCGGGTGCGACGTAAGACGCAAGCCCAAAATGTGAGAGAGATGAGATTGAGGTATGCAAGCTGGAATGTGGGCAGTATGACTGGGAAAGGAAGGGAGCTAGTAGATGTGTTAAAGAGGAGAAGAATAAATATAGCATGTCTGCAAGAGACTAGGTGGAAAGGAGCGAAAGCTAGAGAAATTGGTGAAGGATATAAGCTGTATTATAGTGGAAGTGATGGAAGAAGGAATGGCGTGGGAGTAGTGTTAGATAAGGATTTGAAAGATTGTGTGGCGCATGTCATAAGAACGAGTGATAGGATAATAACCGTAAAAATAGTTTGTGAAAGTGTTATTTTGAATGTTATAAGTGTGTACGCGCCTCAGTCCGGGTGCAAGGAGGACGTGAAAGAAAAATTTTGGCAGGATTTTGATAGTGTTATGATTCGCATTCCAGAGAGTGAAGAGATTTGTATAGGAGGTGATTTTAATGGACATGTAGGAGCTACAAATGAGGGCTATGAAAGGGTGCATGGAGGGTGGGGGTATGGCAATCGTAATGATGATGGAGACCAGTTGCTCCAAGCTGCGACTACCTCTAACCTGGCAGTTGCGAATACGTGGTTCCAGAAACGGCCTGAACACCTTATCACCTATAAGAGTGGTAATCACGCGACACAGATCGACTACTTTCTGATTAAGAGAAGTAAGTTGGTCTGTGTCAAAAACTGCAAAGTACTGCCGGGTGAAGCATTGGTGACGCAGCATAGACTGCTACTAATGGACATTACAATCAGTTACAAGTACCTAAGGAGAAAAAACCGTCTCTCGCCAAAAATTAGATGGCGTATGCTAGAGACAGATGAGTATGCTGGTCGATTTAGGGAAATAATGGTAGAAAACATCTTGGAAATGAAGGATATGAAAGAGAAATCTGCGAATGATTGTTGGGATGAAATGGCAAATAGTGTCAGGAAGACAGCTAAGGCTGTGCTCGGCGAATCAAAAGGGAAAGGTATAATTGACAAGGATACATGGTGGTGGAATGAGAATGTACAGAGAGAATTGAAAGAAAAGAAGAATGCATTCAAAAAGTGGCAACTAGAAAGAGGTAATGAAAGTGAGAGGCAAGCTAGGAAGAATGAGTATAGAGAATGTAAGAAAAAGGCCACTAAAGCGGTTGCTATAGCCAGGTCTGACGCTCAGAAACGGTTGTATGCCGCTTTAGATGGACCTCGTGGACAAAAGGAACTTTACCGTATTACAAGAGCTAGAGAAGAAAGAGCGATATTAAGCATGTCAGATGAGAACGACTGGAATAGAGTGATTAATAGGAAGCCAGAGAACGTCGGACTTGTGAATGAGATTAGTATGTATGAAGTTAGAGAAGCAGTGAGAAGTATGAAAAGCGGAAAATCGGAAGGACCAGACGGTATACCAGTGGAAGTATGGAAGATACTGGGAGAAGACGGATATAAGTGGCTGACTTTATTCTTCAATAAGCTGCTGCAAGAAGAAGTGATCCCTACGGAATGGAGCATCAGTACGCTGGTGCCCATATACAAAAATAAAGGGGATGTGCAAGACTGTGGCAGCTATCGGGGAATAAAGCTTATGTCTCATAGTATGAAAGTTTGGGAGAAAGTGATAGAAAAGCGATTGCGAGATGAAAGTGAGATCACCCAAAACCAGTTCGGGTTCATGCCTGGTCGCGGGACAACGGACGCCATATTTGCACTACGCCAAGTGTGCGAAAAACATCGCGACGTGCACAGGAATCTGCATATGGTGTTCGTTGATCTAGAAAAAGCGTACGACCGAGTACCTAGAGCAGTTTTGTGGTGGGCATTGAATGAGAAAGGTATACCTGGTAAGTATGTGAGGTTAATCTGTGCCATGTACAGTCGAGCCAGGACGTATGTACGAACCGCCGCGGGGAATTCCGACGAGTTCAGTGTGGCAGTGGGCTTACACCAAGGGTCTGCACTAAGTCCCTATCTCTTCCTGCTTGTGATGGATGCCTTGACATCGGAGATACAGGAAGAGCCCCCTTGGTGTATGCTGTTTGCCGATGACATAGTGCTCGTCGGAGAAAACGGGCTCGAGGTCCAAAACATACTGGAGAAATGGCGATGCAAGTTGGAGAGTGTTGGCCTAAAAATCAGCAGATCGAAAACCGAACATCTGTTCTGTGATTTTGGCGGTCTCTCCAATTTTACACCCATTTCCCTCGACGGAACACCTTTGCCAGTATGTCAAGACTTCCGATACCTAGGGTCTGTTATCCAAAGCGACGGTGAACTGGATCGTACTGTGAGGCACAGAATTGACGCAGGATGGATGAAATGGCGGCAGGTCACGGGCACCATATGTGACTCGCACATCCCTCTTCCCCTAAAAGGGAAGATATATAAGACCTTAATAAGGCCTGCCGTCTTGTATGGATCAGCTTGTTGGACAACGAAAGTGGCGGATGAAAGGCGATTGCATGCAGCAGAGATGCGAATGTTGCGATGGATGTGTGGAGTAACGAGAATGGATAGAATACGGAATGAATATGTTAGAGGAAGTCTGAAAGTGGCACCTGTGACAGAGAAGCTGAGGAGTGCACGTTTGGGATGGTATGGACATGTGATGAGACGGAATGAAAATGAGGTTGTTAAGAGAGTGTTAACTATGAATGTGGAAGGATTTAGAGGAAGAGGTAGACCTAAGAAGAAATGA
Protein
MRRNKKKSTRARASPAGDAQGQHPVSVESGRGLSHHGRVRRKTQAQNVREMRLRYASWNVGSMTGKGRELVDVLKRRRINIACLQETRWKGAKAREIGEGYKLYYSGSDGRRNGVGVVLDKDLKDCVAHVIRTSDRIITVKIVCESVILNVISVYAPQSGCKEDVKEKFWQDFDSVMIRIPESEEICIGGDFNGHVGATNEGYERVHGGWGYGNRNDDGDQLLQAATTSNLAVANTWFQKRPEHLITYKSGNHATQIDYFLIKRSKLVCVKNCKVLPGEALVTQHRLLLMDITISYKYLRRKNRLSPKIRWRMLETDEYAGRFREIMVENILEMKDMKEKSANDCWDEMANSVRKTAKAVLGESKGKGIIDKDTWWWNENVQRELKEKKNAFKKWQLERGNESERQARKNEYRECKKKATKAVAIARSDAQKRLYAALDGPRGQKELYRITRAREERAILSMSDENDWNRVINRKPENVGLVNEISMYEVREAVRSMKSGKSEGPDGIPVEVWKILGEDGYKWLTLFFNKLLQEEVIPTEWSISTLVPIYKNKGDVQDCGSYRGIKLMSHSMKVWEKVIEKRLRDESEITQNQFGFMPGRGTTDAIFALRQVCEKHRDVHRNLHMVFVDLEKAYDRVPRAVLWWALNEKGIPGKYVRLICAMYSRARTYVRTAAGNSDEFSVAVGLHQGSALSPYLFLLVMDALTSEIQEEPPWCMLFADDIVLVGENGLEVQNILEKWRCKLESVGLKISRSKTEHLFCDFGGLSNFTPISLDGTPLPVCQDFRYLGSVIQSDGELDRTVRHRIDAGWMKWRQVTGTICDSHIPLPLKGKIYKTLIRPAVLYGSACWTTKVADERRLHAAEMRMLRWMCGVTRMDRIRNEYVRGSLKVAPVTEKLRSARLGWYGHVMRRNENEVVKRVLTMNVEGFRGRGRPKKK
Summary
Uniprot
A0A2A4IZ11
A0A445J0R5
C6Y4D5
A0A445G8Y2
A0A445LEB4
A0A445FNJ6
+ More
A0A445F6Z2 A0A445G8F3 A0A1D6FJU8 A0A1D6M7A6 A0A3L6FAM2 A0A1D6P0V1 A0A1D6K3M7 A0A3L6FHK1 A0A1D6JQ10 A0A1D6HX90 A0A1D6KLY6 A0A3L6GA57 A0A1D6JJN9 A0A1D6I351 A0A445I5D1 A0A1D6I7A1 A0A3L6EF28 A0A3L6DTJ3 A0A1D6LXZ4 A0A1D6G503 A0A445I5C8 A0A1D6I607 A0A445I5E3 A0A1D6KVT7 A0A1D6MV72 A0A1D6J5H5 A0A1D6NCE0 A0A1D6LFI2 A0A445L940 A0A1D6GRX4 A0A3L6G913 A0A1D6PU65 A0A445G821 A0A1D6HE72 A0A445FDA8 A0A3B3C7H7 A0A317YB76 A0A3B3BTI6 A0A3B3CQH3 A0A3B3CTC7 A0A3B3CPF5 A0A3B3E183 A0A3B3BT53 A0A3B3HS69 A0A3B3BC11 A0A0V0IX54 A0A1D6MYV7 A0A3B3CCF8 A0A3Q1I6K9 A0A3B3IEV4 A0A3P9KU10 A0A3B3BH81 A0A3B3BQU5 A0A3P9KEZ0 A0A3B3IC09 A0A3P9M4S0 A0A3P9M3S5 A0A3B3HT45 A0A3B3HJZ3 A0A3Q1JCX2 A0A3P9J281 A0A3B3HKV3 A0A3B3HPV6 A0A3P9H708 A0A3P9MAU2 A0A3B3CWT9 A0A3Q1HIZ5 A0A3B3CWZ8 A0A3B3HQW9 A0A3B3H3Q1 A0A3B3HY61 A0A3B3HLA9 A0A3B3HQU7 A0A3B3IM62 A0A3P9K1J1 A0A3B3HEZ2 A0A3P9K813 A0A3B3H5C8 A0A3B3C2Z2 A0A3P9LQU2 A0A3P9MI82 A0A3B3HFD6 A0A3B3IP13 A0A3P9M241 A0A317Y8S7 A0A1D6KWI6 A0A3Q1HXD9 A0A3B3HUW2 A0A3P9HVA4 A0A3B3ILX4 A0A3P9JFB7 A0A3B3HV05 A0A3B3HVG5 A0A3P9JXW8
A0A445F6Z2 A0A445G8F3 A0A1D6FJU8 A0A1D6M7A6 A0A3L6FAM2 A0A1D6P0V1 A0A1D6K3M7 A0A3L6FHK1 A0A1D6JQ10 A0A1D6HX90 A0A1D6KLY6 A0A3L6GA57 A0A1D6JJN9 A0A1D6I351 A0A445I5D1 A0A1D6I7A1 A0A3L6EF28 A0A3L6DTJ3 A0A1D6LXZ4 A0A1D6G503 A0A445I5C8 A0A1D6I607 A0A445I5E3 A0A1D6KVT7 A0A1D6MV72 A0A1D6J5H5 A0A1D6NCE0 A0A1D6LFI2 A0A445L940 A0A1D6GRX4 A0A3L6G913 A0A1D6PU65 A0A445G821 A0A1D6HE72 A0A445FDA8 A0A3B3C7H7 A0A317YB76 A0A3B3BTI6 A0A3B3CQH3 A0A3B3CTC7 A0A3B3CPF5 A0A3B3E183 A0A3B3BT53 A0A3B3HS69 A0A3B3BC11 A0A0V0IX54 A0A1D6MYV7 A0A3B3CCF8 A0A3Q1I6K9 A0A3B3IEV4 A0A3P9KU10 A0A3B3BH81 A0A3B3BQU5 A0A3P9KEZ0 A0A3B3IC09 A0A3P9M4S0 A0A3P9M3S5 A0A3B3HT45 A0A3B3HJZ3 A0A3Q1JCX2 A0A3P9J281 A0A3B3HKV3 A0A3B3HPV6 A0A3P9H708 A0A3P9MAU2 A0A3B3CWT9 A0A3Q1HIZ5 A0A3B3CWZ8 A0A3B3HQW9 A0A3B3H3Q1 A0A3B3HY61 A0A3B3HLA9 A0A3B3HQU7 A0A3B3IM62 A0A3P9K1J1 A0A3B3HEZ2 A0A3P9K813 A0A3B3H5C8 A0A3B3C2Z2 A0A3P9LQU2 A0A3P9MI82 A0A3B3HFD6 A0A3B3IP13 A0A3P9M241 A0A317Y8S7 A0A1D6KWI6 A0A3Q1HXD9 A0A3B3HUW2 A0A3P9HVA4 A0A3B3ILX4 A0A3P9JFB7 A0A3B3HV05 A0A3B3HVG5 A0A3P9JXW8
EMBL
NWSH01004369
PCG65177.1
QZWG01000009
RZB91994.1
CU462842
CBA11992.1
+ More
QZWG01000017 RZB57624.1 QZWG01000003 RZC21519.1 QZWG01000018 RZB50463.1 QZWG01000020 RZB44550.1 RZB57475.1 CM000784 AQK92036.1 CM000782 AQK86945.1 NCVQ01000004 PWZ29990.1 CM000785 AQL03708.1 CM000780 CM007647 AQK53188.1 ONL98233.1 PWZ32579.1 CM007649 AQK59578.1 ONL94081.1 ONM09892.1 ONM36701.1 CM007650 ONM52836.1 ONM03863.1 NCVQ01000002 PWZ45158.1 ONL92524.1 ONM54584.1 QZWG01000011 RZB81211.1 ONM55942.1 NCVQ01000007 PWZ18651.1 NCVQ01000009 PWZ11925.1 AQK84025.1 AQK98364.1 RZB81213.1 ONM55524.1 RZB81212.1 ONM06616.1 ONM32720.1 CM000786 AQK43188.1 ONM38170.1 AQK78701.1 RZC19618.1 CM000781 AQK65817.1 PWZ44991.1 AQK50169.1 RZB57350.1 RZB57351.1 AQK72949.1 QZWG01000019 RZB46832.1 NCVQ01000001 PWZ54982.1 GEDG01001285 JAP37101.1 ONM33873.1 PWZ54893.1 ONM06831.1
QZWG01000017 RZB57624.1 QZWG01000003 RZC21519.1 QZWG01000018 RZB50463.1 QZWG01000020 RZB44550.1 RZB57475.1 CM000784 AQK92036.1 CM000782 AQK86945.1 NCVQ01000004 PWZ29990.1 CM000785 AQL03708.1 CM000780 CM007647 AQK53188.1 ONL98233.1 PWZ32579.1 CM007649 AQK59578.1 ONL94081.1 ONM09892.1 ONM36701.1 CM007650 ONM52836.1 ONM03863.1 NCVQ01000002 PWZ45158.1 ONL92524.1 ONM54584.1 QZWG01000011 RZB81211.1 ONM55942.1 NCVQ01000007 PWZ18651.1 NCVQ01000009 PWZ11925.1 AQK84025.1 AQK98364.1 RZB81213.1 ONM55524.1 RZB81212.1 ONM06616.1 ONM32720.1 CM000786 AQK43188.1 ONM38170.1 AQK78701.1 RZC19618.1 CM000781 AQK65817.1 PWZ44991.1 AQK50169.1 RZB57350.1 RZB57351.1 AQK72949.1 QZWG01000019 RZB46832.1 NCVQ01000001 PWZ54982.1 GEDG01001285 JAP37101.1 ONM33873.1 PWZ54893.1 ONM06831.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR002495 Glyco_trans_8
IPR029044 Nucleotide-diphossugar_trans
IPR027124 Swc5/CFDP2
IPR011256 Reg_factor_effector_dom_sf
IPR006917 SOUL_haem-bd
IPR036565 Mur-like_cat_sf
IPR001645 Folylpolyglutamate_synth
IPR018109 Folylpolyglutamate_synth_CS
IPR036615 Mur_ligase_C_dom_sf
IPR003593 AAA+_ATPase
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR036869 J_dom_sf
IPR018253 DnaJ_domain_CS
IPR001623 DnaJ_domain
IPR003323 OTU_dom
IPR013103 RVT_2
IPR038765 Papain-like_cys_pep_sf
IPR024964 CTLH/CRA
IPR006595 CTLH_C
IPR007021 DUF659
IPR012337 RNaseH-like_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR002495 Glyco_trans_8
IPR029044 Nucleotide-diphossugar_trans
IPR027124 Swc5/CFDP2
IPR011256 Reg_factor_effector_dom_sf
IPR006917 SOUL_haem-bd
IPR036565 Mur-like_cat_sf
IPR001645 Folylpolyglutamate_synth
IPR018109 Folylpolyglutamate_synth_CS
IPR036615 Mur_ligase_C_dom_sf
IPR003593 AAA+_ATPase
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR027417 P-loop_NTPase
IPR036869 J_dom_sf
IPR018253 DnaJ_domain_CS
IPR001623 DnaJ_domain
IPR003323 OTU_dom
IPR013103 RVT_2
IPR038765 Papain-like_cys_pep_sf
IPR024964 CTLH/CRA
IPR006595 CTLH_C
IPR007021 DUF659
IPR012337 RNaseH-like_sf
SUPFAM
CDD
ProteinModelPortal
A0A2A4IZ11
A0A445J0R5
C6Y4D5
A0A445G8Y2
A0A445LEB4
A0A445FNJ6
+ More
A0A445F6Z2 A0A445G8F3 A0A1D6FJU8 A0A1D6M7A6 A0A3L6FAM2 A0A1D6P0V1 A0A1D6K3M7 A0A3L6FHK1 A0A1D6JQ10 A0A1D6HX90 A0A1D6KLY6 A0A3L6GA57 A0A1D6JJN9 A0A1D6I351 A0A445I5D1 A0A1D6I7A1 A0A3L6EF28 A0A3L6DTJ3 A0A1D6LXZ4 A0A1D6G503 A0A445I5C8 A0A1D6I607 A0A445I5E3 A0A1D6KVT7 A0A1D6MV72 A0A1D6J5H5 A0A1D6NCE0 A0A1D6LFI2 A0A445L940 A0A1D6GRX4 A0A3L6G913 A0A1D6PU65 A0A445G821 A0A1D6HE72 A0A445FDA8 A0A3B3C7H7 A0A317YB76 A0A3B3BTI6 A0A3B3CQH3 A0A3B3CTC7 A0A3B3CPF5 A0A3B3E183 A0A3B3BT53 A0A3B3HS69 A0A3B3BC11 A0A0V0IX54 A0A1D6MYV7 A0A3B3CCF8 A0A3Q1I6K9 A0A3B3IEV4 A0A3P9KU10 A0A3B3BH81 A0A3B3BQU5 A0A3P9KEZ0 A0A3B3IC09 A0A3P9M4S0 A0A3P9M3S5 A0A3B3HT45 A0A3B3HJZ3 A0A3Q1JCX2 A0A3P9J281 A0A3B3HKV3 A0A3B3HPV6 A0A3P9H708 A0A3P9MAU2 A0A3B3CWT9 A0A3Q1HIZ5 A0A3B3CWZ8 A0A3B3HQW9 A0A3B3H3Q1 A0A3B3HY61 A0A3B3HLA9 A0A3B3HQU7 A0A3B3IM62 A0A3P9K1J1 A0A3B3HEZ2 A0A3P9K813 A0A3B3H5C8 A0A3B3C2Z2 A0A3P9LQU2 A0A3P9MI82 A0A3B3HFD6 A0A3B3IP13 A0A3P9M241 A0A317Y8S7 A0A1D6KWI6 A0A3Q1HXD9 A0A3B3HUW2 A0A3P9HVA4 A0A3B3ILX4 A0A3P9JFB7 A0A3B3HV05 A0A3B3HVG5 A0A3P9JXW8
A0A445F6Z2 A0A445G8F3 A0A1D6FJU8 A0A1D6M7A6 A0A3L6FAM2 A0A1D6P0V1 A0A1D6K3M7 A0A3L6FHK1 A0A1D6JQ10 A0A1D6HX90 A0A1D6KLY6 A0A3L6GA57 A0A1D6JJN9 A0A1D6I351 A0A445I5D1 A0A1D6I7A1 A0A3L6EF28 A0A3L6DTJ3 A0A1D6LXZ4 A0A1D6G503 A0A445I5C8 A0A1D6I607 A0A445I5E3 A0A1D6KVT7 A0A1D6MV72 A0A1D6J5H5 A0A1D6NCE0 A0A1D6LFI2 A0A445L940 A0A1D6GRX4 A0A3L6G913 A0A1D6PU65 A0A445G821 A0A1D6HE72 A0A445FDA8 A0A3B3C7H7 A0A317YB76 A0A3B3BTI6 A0A3B3CQH3 A0A3B3CTC7 A0A3B3CPF5 A0A3B3E183 A0A3B3BT53 A0A3B3HS69 A0A3B3BC11 A0A0V0IX54 A0A1D6MYV7 A0A3B3CCF8 A0A3Q1I6K9 A0A3B3IEV4 A0A3P9KU10 A0A3B3BH81 A0A3B3BQU5 A0A3P9KEZ0 A0A3B3IC09 A0A3P9M4S0 A0A3P9M3S5 A0A3B3HT45 A0A3B3HJZ3 A0A3Q1JCX2 A0A3P9J281 A0A3B3HKV3 A0A3B3HPV6 A0A3P9H708 A0A3P9MAU2 A0A3B3CWT9 A0A3Q1HIZ5 A0A3B3CWZ8 A0A3B3HQW9 A0A3B3H3Q1 A0A3B3HY61 A0A3B3HLA9 A0A3B3HQU7 A0A3B3IM62 A0A3P9K1J1 A0A3B3HEZ2 A0A3P9K813 A0A3B3H5C8 A0A3B3C2Z2 A0A3P9LQU2 A0A3P9MI82 A0A3B3HFD6 A0A3B3IP13 A0A3P9M241 A0A317Y8S7 A0A1D6KWI6 A0A3Q1HXD9 A0A3B3HUW2 A0A3P9HVA4 A0A3B3ILX4 A0A3P9JFB7 A0A3B3HV05 A0A3B3HVG5 A0A3P9JXW8
PDB
2VOA
E-value=0.000537209,
Score=106
Ontologies
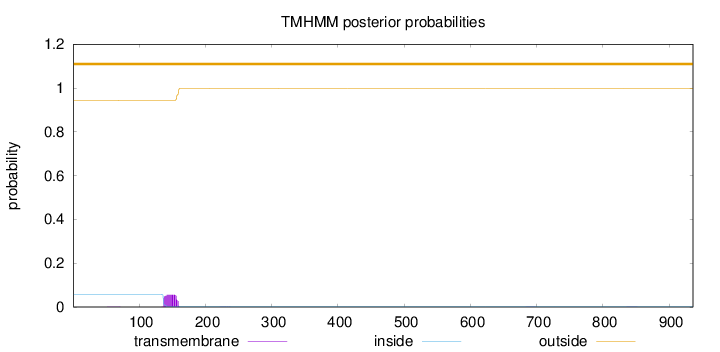
Topology
Length:
936
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.18849
Exp number, first 60 AAs:
0.00256
Total prob of N-in:
0.05731
outside
1 - 936
Population Genetic Test Statistics
Pi
194.821726
Theta
189.429443
Tajima's D
0.557933
CLR
0.42936
CSRT
0.532373381330933
Interpretation
Uncertain
