Gene
KWMTBOMO03794 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010016
Annotation
PREDICTED:_putative_peptidyl-prolyl_cis-trans_isomerase_dodo_isoform_X2_[Amyelois_transitella]
Full name
Peptidyl-prolyl cis-trans isomerase
+ More
Putative peptidyl-prolyl cis-trans isomerase dodo
Putative peptidyl-prolyl cis-trans isomerase dodo
Alternative Name
Rotamase dodo
Location in the cell
Nuclear Reliability : 2.554
Sequence
CDS
ATGTCGAACGAGAATGAACCACCACTGCCCGATGGATGGGAAATGAGAACCAGCCGATCAACAGGAATGACTTACTATCTCAACACTTATACCAAGAAGTCTCAATGGGAGCGTCCTGAAGCACCGGCAGATGCTGGTGAGGTCCGCTGCAGTCATATACTGGTTAAACACGCTGAAAGCCGTCGCCCAACTTCCTGGCGCGAAGAGAAGATCACGCGCACAAAGGAAGAGGCTTTGGAGCTAATCAAAGGCTATCGCAAACAGATTGTTGCAAATGACGCACAGTTTGATGAAATTGCCCTCAAGTATTCTGACTGCTCGTCTGCTAAGCGTGGTGGTGACCTGGGTATGTTTGGGAAAGGACAGACTCAGCTAGCTTTCGAAGAAGAATCTTTTAAGCTGAAAATCGGGCAGTTGAGCAAACCCATAGAGACTGAATCTGGATTACACATCATCCTCCGCACTGCTTAG
Protein
MSNENEPPLPDGWEMRTSRSTGMTYYLNTYTKKSQWERPEAPADAGEVRCSHILVKHAESRRPTSWREEKITRTKEEALELIKGYRKQIVANDAQFDEIALKYSDCSSAKRGGDLGMFGKGQTQLAFEEESFKLKIGQLSKPIETESGLHIILRTA
Summary
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Keywords
Complete proteome
Isomerase
Reference proteome
Rotamase
Feature
chain Putative peptidyl-prolyl cis-trans isomerase dodo
Uniprot
H9JKG5
S4PT88
A0A2H1V2T2
A0A2A4KAN6
A0A0L7LI30
A0A212F7T3
+ More
A0A0N1INT5 I4DNB7 A0A3S2NAD3 A0A1L8EDG8 A0A1I8PTZ2 A0A1I8MFS6 A0A212ETJ5 A0A1A9UHS2 Q2F5X0 A0A154P9S5 A0A224Z4U4 A0A131YJ33 A0A1S3JNA8 A0A0L0C829 A0A1A9WZ95 A0A1B0AIX7 S4NP25 A0A2P8YAL7 A0A2A3EBE2 V9IK58 K7J9W2 A0A1A9XJR9 A0A1B0B754 A0A026WNE1 E0W1N7 A0A067REH8 A0A0M9A459 A0A1B6JFZ9 A0A1B6FHV8 A0A1B6MER9 A0A088A2A7 A0A034VPR7 A0A0J7KLV6 A0A0K8UUG1 A0A3S2NRA2 A0A232EPW9 F4WK86 A0A195BGM2 A0A158NID6 J3JX40 E1ZX16 D6WFG9 A0A3M7QQM9 A0A1W4XGV6 A0A182PAK2 K1QK10 A0A182TMT6 W8C364 A0A348G6B9 A0A182IQ20 A0A131XGA9 L7MCW2 A0A182VPG4 A0A182X8S8 A0A182ID11 A0A146LJK4 B4L4B1 A0A182K362 A0A182XWS9 A0A1L8DU44 A0A1B6ECK2 A0A2H8TYZ8 A0A2S2PKB6 A0A023EG17 T1E8Q9 C4WY86 Q16UF6 Q7QAB6 A0A0A9WM97 A0A182QYX6 A0A1S4FNP2 A0A182R5P2 W5JEQ9 B3NYD4 A0A2M4AXJ4 A0A2M4C1T7 A0A0U2T4T4 A0A0C9S0V3 A0A1Q3F434 B0X6Q0 A0A182VY31 A0A3S3RQX2 A0A2M4AVS0 A0A1Y1KVY6 A0A1S3CTH5 A0A182MJ68 A0A0K8TPM7 A0A023GII1 B4PZ30 A0A0N8ENB8 B4R3R1 A0A2M3Z9W0 A0A411G6R6 P54353
A0A0N1INT5 I4DNB7 A0A3S2NAD3 A0A1L8EDG8 A0A1I8PTZ2 A0A1I8MFS6 A0A212ETJ5 A0A1A9UHS2 Q2F5X0 A0A154P9S5 A0A224Z4U4 A0A131YJ33 A0A1S3JNA8 A0A0L0C829 A0A1A9WZ95 A0A1B0AIX7 S4NP25 A0A2P8YAL7 A0A2A3EBE2 V9IK58 K7J9W2 A0A1A9XJR9 A0A1B0B754 A0A026WNE1 E0W1N7 A0A067REH8 A0A0M9A459 A0A1B6JFZ9 A0A1B6FHV8 A0A1B6MER9 A0A088A2A7 A0A034VPR7 A0A0J7KLV6 A0A0K8UUG1 A0A3S2NRA2 A0A232EPW9 F4WK86 A0A195BGM2 A0A158NID6 J3JX40 E1ZX16 D6WFG9 A0A3M7QQM9 A0A1W4XGV6 A0A182PAK2 K1QK10 A0A182TMT6 W8C364 A0A348G6B9 A0A182IQ20 A0A131XGA9 L7MCW2 A0A182VPG4 A0A182X8S8 A0A182ID11 A0A146LJK4 B4L4B1 A0A182K362 A0A182XWS9 A0A1L8DU44 A0A1B6ECK2 A0A2H8TYZ8 A0A2S2PKB6 A0A023EG17 T1E8Q9 C4WY86 Q16UF6 Q7QAB6 A0A0A9WM97 A0A182QYX6 A0A1S4FNP2 A0A182R5P2 W5JEQ9 B3NYD4 A0A2M4AXJ4 A0A2M4C1T7 A0A0U2T4T4 A0A0C9S0V3 A0A1Q3F434 B0X6Q0 A0A182VY31 A0A3S3RQX2 A0A2M4AVS0 A0A1Y1KVY6 A0A1S3CTH5 A0A182MJ68 A0A0K8TPM7 A0A023GII1 B4PZ30 A0A0N8ENB8 B4R3R1 A0A2M3Z9W0 A0A411G6R6 P54353
EC Number
5.2.1.8
Pubmed
19121390
23622113
26227816
22118469
26354079
22651552
+ More
25315136 28797301 26830274 26108605 29403074 20075255 24508170 30249741 20566863 24845553 25348373 28648823 21719571 21347285 22516182 20798317 18362917 19820115 30375419 22992520 24495485 28049606 25576852 26823975 17994087 25244985 24945155 17510324 12364791 14747013 17210077 25401762 20920257 23761445 26621068 26131772 28004739 26369729 17550304 8552658 9520435 10731132 12537572
25315136 28797301 26830274 26108605 29403074 20075255 24508170 30249741 20566863 24845553 25348373 28648823 21719571 21347285 22516182 20798317 18362917 19820115 30375419 22992520 24495485 28049606 25576852 26823975 17994087 25244985 24945155 17510324 12364791 14747013 17210077 25401762 20920257 23761445 26621068 26131772 28004739 26369729 17550304 8552658 9520435 10731132 12537572
EMBL
BABH01024778
GAIX01012463
JAA80097.1
ODYU01000417
SOQ35155.1
NWSH01000006
+ More
PCG80974.1 JTDY01001097 KOB74886.1 AGBW02009831 OWR49794.1 KQ460869 KPJ11798.1 AK402827 KQ459167 BAM19407.1 KPJ03480.1 RSAL01000151 RVE45745.1 GFDG01002087 JAV16712.1 AGBW02012593 OWR44771.1 DQ311303 ABD36247.1 KQ434844 KZC08144.1 GFPF01010457 MAA21603.1 GEDV01010471 JAP78086.1 JRES01000777 KNC28416.1 GAIX01012024 JAA80536.1 PYGN01000753 PSN41302.1 KZ288311 PBC28529.1 JR048481 AEY60679.1 AAZX01013758 JXJN01009403 KK107145 QOIP01000010 EZA57493.1 RLU18172.1 DS235873 EEB19619.1 KK852685 KDR18483.1 KQ435762 KOX75489.1 GECU01009599 JAS98107.1 GECZ01020153 GECZ01019981 JAS49616.1 JAS49788.1 GEBQ01005547 JAT34430.1 GAKP01015152 GAKP01015148 GAKP01015144 JAC43800.1 LBMM01005784 KMQ91206.1 GDHF01022324 JAI29990.1 RSAL01000006 RVE54317.1 NNAY01002872 OXU20400.1 GL888197 EGI65389.1 KQ976491 KYM83320.1 ADTU01016582 BT127808 AEE62770.1 GL434950 EFN74266.1 KQ971327 EFA00919.1 REGN01005431 RNA13391.1 JH817521 EKC34118.1 GAMC01010298 GAMC01010296 JAB96259.1 FX986035 BBF97992.1 GEFH01003376 JAP65205.1 GACK01002988 JAA62046.1 APCN01004963 GDHC01020926 GDHC01020022 GDHC01011154 GDHC01004781 JAP97702.1 JAP98606.1 JAQ07475.1 JAQ13848.1 CH933810 EDW07389.1 GFDF01004150 JAV09934.1 GEDC01001647 JAS35651.1 GFXV01007314 MBW19119.1 GGMR01017283 MBY29902.1 GAPW01005311 JAC08287.1 GAMD01002553 JAA99037.1 ABLF02040575 AK343080 BAH72856.1 CH477622 EAT38181.1 AAAB01008898 EAA09299.3 GBHO01043834 GBHO01043831 GBHO01043823 GBHO01042783 GBHO01042780 GBHO01037614 GBHO01026212 GBHO01016485 GBHO01016484 GBHO01016483 GBHO01007795 GBRD01008327 GBRD01006312 JAF99769.1 JAF99772.1 JAF99780.1 JAG00821.1 JAG00824.1 JAG05990.1 JAG17392.1 JAG27119.1 JAG27120.1 JAG27121.1 JAG35809.1 JAG57494.1 AXCN02001168 ADMH02001607 ETN61828.1 CH954180 EDV47613.1 GGFK01012183 MBW45504.1 GGFJ01010131 MBW59272.1 KT754663 ALS04497.1 GBZX01002391 JAG90349.1 GFDL01012719 JAV22326.1 DS232420 EDS41533.1 NCKU01006630 NCKU01006558 NCKU01001938 RWS03305.1 RWS03382.1 RWS10856.1 GGFK01011491 MBW44812.1 GEZM01074164 JAV64651.1 AXCM01006361 GDAI01001753 JAI15850.1 GBBM01002858 JAC32560.1 CM000162 EDX03091.1 GDIQ01009924 JAN84813.1 CM000366 EDX18529.1 GGFM01004576 MBW25327.1 MH365800 QBB01609.1 U35140 AF017777 AE014298 BT099557
PCG80974.1 JTDY01001097 KOB74886.1 AGBW02009831 OWR49794.1 KQ460869 KPJ11798.1 AK402827 KQ459167 BAM19407.1 KPJ03480.1 RSAL01000151 RVE45745.1 GFDG01002087 JAV16712.1 AGBW02012593 OWR44771.1 DQ311303 ABD36247.1 KQ434844 KZC08144.1 GFPF01010457 MAA21603.1 GEDV01010471 JAP78086.1 JRES01000777 KNC28416.1 GAIX01012024 JAA80536.1 PYGN01000753 PSN41302.1 KZ288311 PBC28529.1 JR048481 AEY60679.1 AAZX01013758 JXJN01009403 KK107145 QOIP01000010 EZA57493.1 RLU18172.1 DS235873 EEB19619.1 KK852685 KDR18483.1 KQ435762 KOX75489.1 GECU01009599 JAS98107.1 GECZ01020153 GECZ01019981 JAS49616.1 JAS49788.1 GEBQ01005547 JAT34430.1 GAKP01015152 GAKP01015148 GAKP01015144 JAC43800.1 LBMM01005784 KMQ91206.1 GDHF01022324 JAI29990.1 RSAL01000006 RVE54317.1 NNAY01002872 OXU20400.1 GL888197 EGI65389.1 KQ976491 KYM83320.1 ADTU01016582 BT127808 AEE62770.1 GL434950 EFN74266.1 KQ971327 EFA00919.1 REGN01005431 RNA13391.1 JH817521 EKC34118.1 GAMC01010298 GAMC01010296 JAB96259.1 FX986035 BBF97992.1 GEFH01003376 JAP65205.1 GACK01002988 JAA62046.1 APCN01004963 GDHC01020926 GDHC01020022 GDHC01011154 GDHC01004781 JAP97702.1 JAP98606.1 JAQ07475.1 JAQ13848.1 CH933810 EDW07389.1 GFDF01004150 JAV09934.1 GEDC01001647 JAS35651.1 GFXV01007314 MBW19119.1 GGMR01017283 MBY29902.1 GAPW01005311 JAC08287.1 GAMD01002553 JAA99037.1 ABLF02040575 AK343080 BAH72856.1 CH477622 EAT38181.1 AAAB01008898 EAA09299.3 GBHO01043834 GBHO01043831 GBHO01043823 GBHO01042783 GBHO01042780 GBHO01037614 GBHO01026212 GBHO01016485 GBHO01016484 GBHO01016483 GBHO01007795 GBRD01008327 GBRD01006312 JAF99769.1 JAF99772.1 JAF99780.1 JAG00821.1 JAG00824.1 JAG05990.1 JAG17392.1 JAG27119.1 JAG27120.1 JAG27121.1 JAG35809.1 JAG57494.1 AXCN02001168 ADMH02001607 ETN61828.1 CH954180 EDV47613.1 GGFK01012183 MBW45504.1 GGFJ01010131 MBW59272.1 KT754663 ALS04497.1 GBZX01002391 JAG90349.1 GFDL01012719 JAV22326.1 DS232420 EDS41533.1 NCKU01006630 NCKU01006558 NCKU01001938 RWS03305.1 RWS03382.1 RWS10856.1 GGFK01011491 MBW44812.1 GEZM01074164 JAV64651.1 AXCM01006361 GDAI01001753 JAI15850.1 GBBM01002858 JAC32560.1 CM000162 EDX03091.1 GDIQ01009924 JAN84813.1 CM000366 EDX18529.1 GGFM01004576 MBW25327.1 MH365800 QBB01609.1 U35140 AF017777 AE014298 BT099557
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000053268
+ More
UP000283053 UP000095300 UP000095301 UP000078200 UP000076502 UP000085678 UP000037069 UP000091820 UP000092445 UP000245037 UP000242457 UP000002358 UP000092443 UP000092460 UP000053097 UP000279307 UP000009046 UP000027135 UP000053105 UP000005203 UP000036403 UP000215335 UP000007755 UP000078540 UP000005205 UP000000311 UP000007266 UP000276133 UP000192223 UP000075885 UP000005408 UP000075902 UP000075880 UP000075903 UP000076407 UP000075840 UP000009192 UP000075881 UP000076408 UP000007819 UP000008820 UP000007062 UP000075886 UP000075900 UP000000673 UP000008711 UP000002320 UP000075920 UP000285301 UP000079169 UP000075883 UP000002282 UP000000304 UP000000803
UP000283053 UP000095300 UP000095301 UP000078200 UP000076502 UP000085678 UP000037069 UP000091820 UP000092445 UP000245037 UP000242457 UP000002358 UP000092443 UP000092460 UP000053097 UP000279307 UP000009046 UP000027135 UP000053105 UP000005203 UP000036403 UP000215335 UP000007755 UP000078540 UP000005205 UP000000311 UP000007266 UP000276133 UP000192223 UP000075885 UP000005408 UP000075902 UP000075880 UP000075903 UP000076407 UP000075840 UP000009192 UP000075881 UP000076408 UP000007819 UP000008820 UP000007062 UP000075886 UP000075900 UP000000673 UP000008711 UP000002320 UP000075920 UP000285301 UP000079169 UP000075883 UP000002282 UP000000304 UP000000803
SUPFAM
SSF51045
SSF51045
CDD
ProteinModelPortal
H9JKG5
S4PT88
A0A2H1V2T2
A0A2A4KAN6
A0A0L7LI30
A0A212F7T3
+ More
A0A0N1INT5 I4DNB7 A0A3S2NAD3 A0A1L8EDG8 A0A1I8PTZ2 A0A1I8MFS6 A0A212ETJ5 A0A1A9UHS2 Q2F5X0 A0A154P9S5 A0A224Z4U4 A0A131YJ33 A0A1S3JNA8 A0A0L0C829 A0A1A9WZ95 A0A1B0AIX7 S4NP25 A0A2P8YAL7 A0A2A3EBE2 V9IK58 K7J9W2 A0A1A9XJR9 A0A1B0B754 A0A026WNE1 E0W1N7 A0A067REH8 A0A0M9A459 A0A1B6JFZ9 A0A1B6FHV8 A0A1B6MER9 A0A088A2A7 A0A034VPR7 A0A0J7KLV6 A0A0K8UUG1 A0A3S2NRA2 A0A232EPW9 F4WK86 A0A195BGM2 A0A158NID6 J3JX40 E1ZX16 D6WFG9 A0A3M7QQM9 A0A1W4XGV6 A0A182PAK2 K1QK10 A0A182TMT6 W8C364 A0A348G6B9 A0A182IQ20 A0A131XGA9 L7MCW2 A0A182VPG4 A0A182X8S8 A0A182ID11 A0A146LJK4 B4L4B1 A0A182K362 A0A182XWS9 A0A1L8DU44 A0A1B6ECK2 A0A2H8TYZ8 A0A2S2PKB6 A0A023EG17 T1E8Q9 C4WY86 Q16UF6 Q7QAB6 A0A0A9WM97 A0A182QYX6 A0A1S4FNP2 A0A182R5P2 W5JEQ9 B3NYD4 A0A2M4AXJ4 A0A2M4C1T7 A0A0U2T4T4 A0A0C9S0V3 A0A1Q3F434 B0X6Q0 A0A182VY31 A0A3S3RQX2 A0A2M4AVS0 A0A1Y1KVY6 A0A1S3CTH5 A0A182MJ68 A0A0K8TPM7 A0A023GII1 B4PZ30 A0A0N8ENB8 B4R3R1 A0A2M3Z9W0 A0A411G6R6 P54353
A0A0N1INT5 I4DNB7 A0A3S2NAD3 A0A1L8EDG8 A0A1I8PTZ2 A0A1I8MFS6 A0A212ETJ5 A0A1A9UHS2 Q2F5X0 A0A154P9S5 A0A224Z4U4 A0A131YJ33 A0A1S3JNA8 A0A0L0C829 A0A1A9WZ95 A0A1B0AIX7 S4NP25 A0A2P8YAL7 A0A2A3EBE2 V9IK58 K7J9W2 A0A1A9XJR9 A0A1B0B754 A0A026WNE1 E0W1N7 A0A067REH8 A0A0M9A459 A0A1B6JFZ9 A0A1B6FHV8 A0A1B6MER9 A0A088A2A7 A0A034VPR7 A0A0J7KLV6 A0A0K8UUG1 A0A3S2NRA2 A0A232EPW9 F4WK86 A0A195BGM2 A0A158NID6 J3JX40 E1ZX16 D6WFG9 A0A3M7QQM9 A0A1W4XGV6 A0A182PAK2 K1QK10 A0A182TMT6 W8C364 A0A348G6B9 A0A182IQ20 A0A131XGA9 L7MCW2 A0A182VPG4 A0A182X8S8 A0A182ID11 A0A146LJK4 B4L4B1 A0A182K362 A0A182XWS9 A0A1L8DU44 A0A1B6ECK2 A0A2H8TYZ8 A0A2S2PKB6 A0A023EG17 T1E8Q9 C4WY86 Q16UF6 Q7QAB6 A0A0A9WM97 A0A182QYX6 A0A1S4FNP2 A0A182R5P2 W5JEQ9 B3NYD4 A0A2M4AXJ4 A0A2M4C1T7 A0A0U2T4T4 A0A0C9S0V3 A0A1Q3F434 B0X6Q0 A0A182VY31 A0A3S3RQX2 A0A2M4AVS0 A0A1Y1KVY6 A0A1S3CTH5 A0A182MJ68 A0A0K8TPM7 A0A023GII1 B4PZ30 A0A0N8ENB8 B4R3R1 A0A2M3Z9W0 A0A411G6R6 P54353
PDB
1F8A
E-value=9.34286e-43,
Score=431
Ontologies
GO
Topology
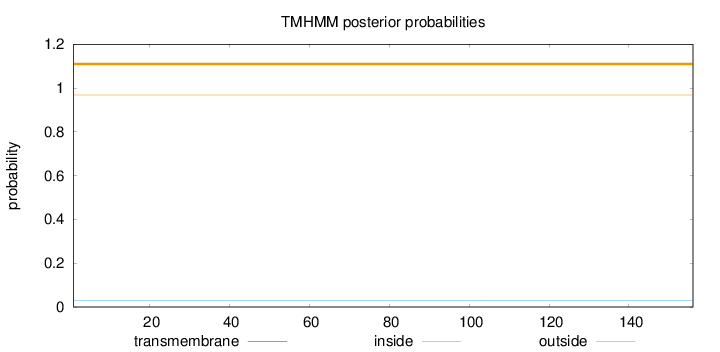
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00048
Exp number, first 60 AAs:
0.00048
Total prob of N-in:
0.03127
outside
1 - 156
Population Genetic Test Statistics
Pi
13.685942
Theta
26.394164
Tajima's D
-1.393375
CLR
0
CSRT
0.0732463376831159
Interpretation
Uncertain
