Gene
KWMTBOMO03791
Pre Gene Modal
BGIBMGA010074
Annotation
hypothetical_protein_KGM_05503_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.263
Sequence
CDS
ATGCATATCCTCTGTACTATATGCAGCGACCTAATAAATCAATCTGAAAGTATATATGTGACAAAATGCGGTCACATATTTCATCATCACTGCCTTATACAGTGGATTGAAAGGTCAAAATCCTGCCCGCAATGCCGCAACAAAGTCACAGAGAAATGCATGTTCCGAATGTATCCTACAATATCAAATGATGTGACAAGCGGCGAAGACTCAGCTACTTTACAGTCTAGATTGGATGACACACAATTACAGCTCAGGCAACAAAAATCTGTGTGCAGAGATAAAGAGGACAAAATATCTGATTTAATAGGGGAGCTAAAAAAGAATGAAGCTCTATTAAAATCATATGAAAAGAAGTTGGTAAGCAGAGATTCTGCAATGACTGCTATGAGAGAACAAATGGAGTACCTCAAAATTCAAAACAAAGAGACAGTCAAGTTAAAAGAGGAAAATGAAACAATGAAGCAGAATTTACAATTGTTAAATGGTTTAAATAACATATTAAATGCAACATGTGAAGACGTAGAGAAGATGCTCCAAGGATACACAGATGTTAAAACCGTTGCTATATTTGCTACAGCGCTTAAAAGAGGTCTCTGCGAGTCGGAAGCCAAGAAGCGCGAGTCGCGCGACAAGCTCCACGCAGCCAAGCAGCAGATCGCAGCCGAGAAGCTCAACGTAAGGGACTTGCAGGCCAAGTTGCTCCATGCAGAAGATAAACTGACGGACATGGAAAGGAAATATCAAGCACTGGCCAAAAGGAAAGCCGGAGATATGACCGAGAAATGTATTGATGACAAGCCGGCTCACGCGGAGGCGCACTCTCTCAAGCACATGCGGCCGAGCTTGGAGAACGTTGAATCTTTAAATACCACTAATAATACCAGCATTAGGACGTTAGTGGACCGTATAGAAAATGCAGATTCGCCGTATCTAAGTCTGAAACAAAGCAATCTGGCTCTAACTGCGTTGCAGAGGGCGCCGAAACATGCCCTATCCCTAGACAGGATCAAGCCATCGGAGTTAGCTATAATAAACTCAGCCAGAAGCACTGTCTCGAAGAAGCCATTAGACAATAAATCATTAAGCATATTTGCCAAAAAAGAGCCAGTAACGTTAGATCCAGATGTCGAGATGGAGTCCTCACAGAACAATATCTGCTATGATGGACTCGGAGGGCATTCTAAATTGGAAATATTTCCAGTTCCCAGTAATAGACCTCTGAAGAGCTGTGTGCCCAAAGTCACTCCCAGGCACCGATTAAAAAGACCAGCATCTTCGGGAAGTCAGGATATCGGCAAGATGATTGAGAAGTTAATGGACAAGTGA
Protein
MHILCTICSDLINQSESIYVTKCGHIFHHHCLIQWIERSKSCPQCRNKVTEKCMFRMYPTISNDVTSGEDSATLQSRLDDTQLQLRQQKSVCRDKEDKISDLIGELKKNEALLKSYEKKLVSRDSAMTAMREQMEYLKIQNKETVKLKEENETMKQNLQLLNGLNNILNATCEDVEKMLQGYTDVKTVAIFATALKRGLCESEAKKRESRDKLHAAKQQIAAEKLNVRDLQAKLLHAEDKLTDMERKYQALAKRKAGDMTEKCIDDKPAHAEAHSLKHMRPSLENVESLNTTNNTSIRTLVDRIENADSPYLSLKQSNLALTALQRAPKHALSLDRIKPSELAIINSARSTVSKKPLDNKSLSIFAKKEPVTLDPDVEMESSQNNICYDGLGGHSKLEIFPVPSNRPLKSCVPKVTPRHRLKRPASSGSQDIGKMIEKLMDK
Summary
Uniprot
H9JKM3
A0A3S2NQB5
A0A212F7T4
S4NXF0
A0A2A4IYB5
A0A0N0PBX7
+ More
A0A194QDE0 A0A0N1IE29 A0A067QK59 A0A182JYN0 A0A182LKD2 B4J4L9 A0A182I4J1 A0A2J7PT31 Q7Q568 B0WCA9 W5JVE3 A0A182VJW3 A0A182X4J4 A0A182H2D8 B4LN04 A0A182PMV1 A0A182NI94 A0A1Y1KAB6 B3MBD9 A0A2M4BMG0 A0A084WGJ5 A0A2M4AXV4 A0A182FD18 A0A2A3E884 B4KMP0 A0A2M4BMX5 A0A1Q3F0R5 D6WTZ7 A0A088AJH3 B3NLR0 Q16FI7 A0A154P9P1 A0A1J1I893 Q7K2X1 B4HNK0 B4P4Q1 B4QCA5 A0A0B4KFV0 B4NN27 Q28YU7 B4GIP5 A0A3B0JQG6 A0A2P8YJ82 A0A0N0BBK4 A0A182WBQ4 A0A1W4UYI7 A0A0M5IY38 A0A182MX11 A0A182R961 Q17EK2 A0A336KCN0 A0A336KBD6 A0A1B0GPF3 A0A1I8M9M8 A0A1B0AGX8 A0A182U6G2 A0A1B0GQ69 A0A3L8DXC9 A0A026WQ86 A0A1A9XTW3 A0A1B0B6E9 T1GW77 W8C3A1 A0A034WVK4 A0A0L7REX8 K7J3Y5 A0A0K8UIP3 A0A1W4WZ74 A0A1I8Q809 A0A0C9RNE7 A0A182Y2J6 A0A3Q3QER8 A0A1W4XA34 A0A1B0CFY3 H2UEV8 A0A2D0PJS9 A0A1B6BZ66 W5U909
A0A194QDE0 A0A0N1IE29 A0A067QK59 A0A182JYN0 A0A182LKD2 B4J4L9 A0A182I4J1 A0A2J7PT31 Q7Q568 B0WCA9 W5JVE3 A0A182VJW3 A0A182X4J4 A0A182H2D8 B4LN04 A0A182PMV1 A0A182NI94 A0A1Y1KAB6 B3MBD9 A0A2M4BMG0 A0A084WGJ5 A0A2M4AXV4 A0A182FD18 A0A2A3E884 B4KMP0 A0A2M4BMX5 A0A1Q3F0R5 D6WTZ7 A0A088AJH3 B3NLR0 Q16FI7 A0A154P9P1 A0A1J1I893 Q7K2X1 B4HNK0 B4P4Q1 B4QCA5 A0A0B4KFV0 B4NN27 Q28YU7 B4GIP5 A0A3B0JQG6 A0A2P8YJ82 A0A0N0BBK4 A0A182WBQ4 A0A1W4UYI7 A0A0M5IY38 A0A182MX11 A0A182R961 Q17EK2 A0A336KCN0 A0A336KBD6 A0A1B0GPF3 A0A1I8M9M8 A0A1B0AGX8 A0A182U6G2 A0A1B0GQ69 A0A3L8DXC9 A0A026WQ86 A0A1A9XTW3 A0A1B0B6E9 T1GW77 W8C3A1 A0A034WVK4 A0A0L7REX8 K7J3Y5 A0A0K8UIP3 A0A1W4WZ74 A0A1I8Q809 A0A0C9RNE7 A0A182Y2J6 A0A3Q3QER8 A0A1W4XA34 A0A1B0CFY3 H2UEV8 A0A2D0PJS9 A0A1B6BZ66 W5U909
Pubmed
19121390
22118469
23622113
26354079
24845553
20966253
+ More
17994087 12364791 14747013 17210077 20920257 23761445 26483478 28004739 24438588 18362917 19820115 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 29403074 25315136 30249741 24508170 24495485 25348373 20075255 25244985 21551351 23127152
17994087 12364791 14747013 17210077 20920257 23761445 26483478 28004739 24438588 18362917 19820115 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 15632085 29403074 25315136 30249741 24508170 24495485 25348373 20075255 25244985 21551351 23127152
EMBL
BABH01024771
BABH01024772
BABH01024773
BABH01024774
BABH01024775
RSAL01000151
+ More
RVE45748.1 AGBW02009831 OWR49790.1 GAIX01012202 JAA80358.1 NWSH01005605 PCG64103.1 KQ460869 KPJ11795.1 KQ459167 KPJ03477.1 KQ458791 KPJ05091.1 KK853244 KDR09434.1 CH916367 EDW02724.1 APCN01002315 NEVH01021921 PNF19487.1 AAAB01008960 EAA11334.4 DS231886 EDS43354.1 ADMH02000378 ETN66774.1 JXUM01105104 KQ564939 KXJ71535.1 CH940648 EDW62119.1 GEZM01087893 JAV58413.1 CH902619 EDV36064.1 GGFJ01005126 MBW54267.1 ATLV01023587 KE525344 KFB49339.1 GGFK01012289 MBW45610.1 KZ288349 PBC27416.1 CH933808 EDW08782.1 GGFJ01005150 MBW54291.1 GFDL01013967 JAV21078.1 KQ971352 EFA07352.2 CH954179 EDV55035.1 CH478429 EAT32998.1 KQ434850 KZC08551.1 CVRI01000043 CRK96435.1 AE013599 AY060610 AAF57730.2 AAL28158.1 CH480816 EDW48419.1 CM000158 EDW91674.1 CM000362 CM002911 EDX07629.1 KMY94788.1 AGB93603.1 CH964282 EDW85766.1 CM000071 EAL25867.1 CH479183 EDW36365.1 OUUW01000001 SPP75899.1 PYGN01000555 PSN44320.1 KQ436240 KOX67348.1 CP012524 ALC42581.1 AXCM01001383 CH477282 EAT44877.1 UFQS01000264 UFQT01000264 SSX02197.1 SSX22574.1 SSX02196.1 SSX22573.1 AJVK01033650 AJVK01035956 QOIP01000003 RLU25130.1 KK107139 EZA57861.1 JXJN01009064 CAQQ02153968 CAQQ02153969 CAQQ02153970 GAMC01010251 JAB96304.1 GAKP01000293 JAC58659.1 KQ414606 KOC69532.1 AAZX01001002 GDHF01026109 GDHF01011015 JAI26205.1 JAI41299.1 GBYB01015022 JAG84789.1 AJWK01010557 GEDC01030924 JAS06374.1 JT408259 AHH38285.1
RVE45748.1 AGBW02009831 OWR49790.1 GAIX01012202 JAA80358.1 NWSH01005605 PCG64103.1 KQ460869 KPJ11795.1 KQ459167 KPJ03477.1 KQ458791 KPJ05091.1 KK853244 KDR09434.1 CH916367 EDW02724.1 APCN01002315 NEVH01021921 PNF19487.1 AAAB01008960 EAA11334.4 DS231886 EDS43354.1 ADMH02000378 ETN66774.1 JXUM01105104 KQ564939 KXJ71535.1 CH940648 EDW62119.1 GEZM01087893 JAV58413.1 CH902619 EDV36064.1 GGFJ01005126 MBW54267.1 ATLV01023587 KE525344 KFB49339.1 GGFK01012289 MBW45610.1 KZ288349 PBC27416.1 CH933808 EDW08782.1 GGFJ01005150 MBW54291.1 GFDL01013967 JAV21078.1 KQ971352 EFA07352.2 CH954179 EDV55035.1 CH478429 EAT32998.1 KQ434850 KZC08551.1 CVRI01000043 CRK96435.1 AE013599 AY060610 AAF57730.2 AAL28158.1 CH480816 EDW48419.1 CM000158 EDW91674.1 CM000362 CM002911 EDX07629.1 KMY94788.1 AGB93603.1 CH964282 EDW85766.1 CM000071 EAL25867.1 CH479183 EDW36365.1 OUUW01000001 SPP75899.1 PYGN01000555 PSN44320.1 KQ436240 KOX67348.1 CP012524 ALC42581.1 AXCM01001383 CH477282 EAT44877.1 UFQS01000264 UFQT01000264 SSX02197.1 SSX22574.1 SSX02196.1 SSX22573.1 AJVK01033650 AJVK01035956 QOIP01000003 RLU25130.1 KK107139 EZA57861.1 JXJN01009064 CAQQ02153968 CAQQ02153969 CAQQ02153970 GAMC01010251 JAB96304.1 GAKP01000293 JAC58659.1 KQ414606 KOC69532.1 AAZX01001002 GDHF01026109 GDHF01011015 JAI26205.1 JAI41299.1 GBYB01015022 JAG84789.1 AJWK01010557 GEDC01030924 JAS06374.1 JT408259 AHH38285.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000218220
UP000053240
UP000053268
+ More
UP000027135 UP000075881 UP000075882 UP000001070 UP000075840 UP000235965 UP000007062 UP000002320 UP000000673 UP000075903 UP000076407 UP000069940 UP000249989 UP000008792 UP000075885 UP000075884 UP000007801 UP000030765 UP000069272 UP000242457 UP000009192 UP000007266 UP000005203 UP000008711 UP000008820 UP000076502 UP000183832 UP000000803 UP000001292 UP000002282 UP000000304 UP000007798 UP000001819 UP000008744 UP000268350 UP000245037 UP000053105 UP000075920 UP000192221 UP000092553 UP000075883 UP000075900 UP000092462 UP000095301 UP000092445 UP000075902 UP000279307 UP000053097 UP000092443 UP000092460 UP000015102 UP000053825 UP000002358 UP000192223 UP000095300 UP000076408 UP000261600 UP000092461 UP000005226 UP000221080
UP000027135 UP000075881 UP000075882 UP000001070 UP000075840 UP000235965 UP000007062 UP000002320 UP000000673 UP000075903 UP000076407 UP000069940 UP000249989 UP000008792 UP000075885 UP000075884 UP000007801 UP000030765 UP000069272 UP000242457 UP000009192 UP000007266 UP000005203 UP000008711 UP000008820 UP000076502 UP000183832 UP000000803 UP000001292 UP000002282 UP000000304 UP000007798 UP000001819 UP000008744 UP000268350 UP000245037 UP000053105 UP000075920 UP000192221 UP000092553 UP000075883 UP000075900 UP000092462 UP000095301 UP000092445 UP000075902 UP000279307 UP000053097 UP000092443 UP000092460 UP000015102 UP000053825 UP000002358 UP000192223 UP000095300 UP000076408 UP000261600 UP000092461 UP000005226 UP000221080
Pfam
PF13639 zf-RING_2
Interpro
SUPFAM
SSF51695
SSF51695
Gene 3D
ProteinModelPortal
H9JKM3
A0A3S2NQB5
A0A212F7T4
S4NXF0
A0A2A4IYB5
A0A0N0PBX7
+ More
A0A194QDE0 A0A0N1IE29 A0A067QK59 A0A182JYN0 A0A182LKD2 B4J4L9 A0A182I4J1 A0A2J7PT31 Q7Q568 B0WCA9 W5JVE3 A0A182VJW3 A0A182X4J4 A0A182H2D8 B4LN04 A0A182PMV1 A0A182NI94 A0A1Y1KAB6 B3MBD9 A0A2M4BMG0 A0A084WGJ5 A0A2M4AXV4 A0A182FD18 A0A2A3E884 B4KMP0 A0A2M4BMX5 A0A1Q3F0R5 D6WTZ7 A0A088AJH3 B3NLR0 Q16FI7 A0A154P9P1 A0A1J1I893 Q7K2X1 B4HNK0 B4P4Q1 B4QCA5 A0A0B4KFV0 B4NN27 Q28YU7 B4GIP5 A0A3B0JQG6 A0A2P8YJ82 A0A0N0BBK4 A0A182WBQ4 A0A1W4UYI7 A0A0M5IY38 A0A182MX11 A0A182R961 Q17EK2 A0A336KCN0 A0A336KBD6 A0A1B0GPF3 A0A1I8M9M8 A0A1B0AGX8 A0A182U6G2 A0A1B0GQ69 A0A3L8DXC9 A0A026WQ86 A0A1A9XTW3 A0A1B0B6E9 T1GW77 W8C3A1 A0A034WVK4 A0A0L7REX8 K7J3Y5 A0A0K8UIP3 A0A1W4WZ74 A0A1I8Q809 A0A0C9RNE7 A0A182Y2J6 A0A3Q3QER8 A0A1W4XA34 A0A1B0CFY3 H2UEV8 A0A2D0PJS9 A0A1B6BZ66 W5U909
A0A194QDE0 A0A0N1IE29 A0A067QK59 A0A182JYN0 A0A182LKD2 B4J4L9 A0A182I4J1 A0A2J7PT31 Q7Q568 B0WCA9 W5JVE3 A0A182VJW3 A0A182X4J4 A0A182H2D8 B4LN04 A0A182PMV1 A0A182NI94 A0A1Y1KAB6 B3MBD9 A0A2M4BMG0 A0A084WGJ5 A0A2M4AXV4 A0A182FD18 A0A2A3E884 B4KMP0 A0A2M4BMX5 A0A1Q3F0R5 D6WTZ7 A0A088AJH3 B3NLR0 Q16FI7 A0A154P9P1 A0A1J1I893 Q7K2X1 B4HNK0 B4P4Q1 B4QCA5 A0A0B4KFV0 B4NN27 Q28YU7 B4GIP5 A0A3B0JQG6 A0A2P8YJ82 A0A0N0BBK4 A0A182WBQ4 A0A1W4UYI7 A0A0M5IY38 A0A182MX11 A0A182R961 Q17EK2 A0A336KCN0 A0A336KBD6 A0A1B0GPF3 A0A1I8M9M8 A0A1B0AGX8 A0A182U6G2 A0A1B0GQ69 A0A3L8DXC9 A0A026WQ86 A0A1A9XTW3 A0A1B0B6E9 T1GW77 W8C3A1 A0A034WVK4 A0A0L7REX8 K7J3Y5 A0A0K8UIP3 A0A1W4WZ74 A0A1I8Q809 A0A0C9RNE7 A0A182Y2J6 A0A3Q3QER8 A0A1W4XA34 A0A1B0CFY3 H2UEV8 A0A2D0PJS9 A0A1B6BZ66 W5U909
PDB
2ECT
E-value=1.65142e-05,
Score=116
Ontologies
KEGG
GO
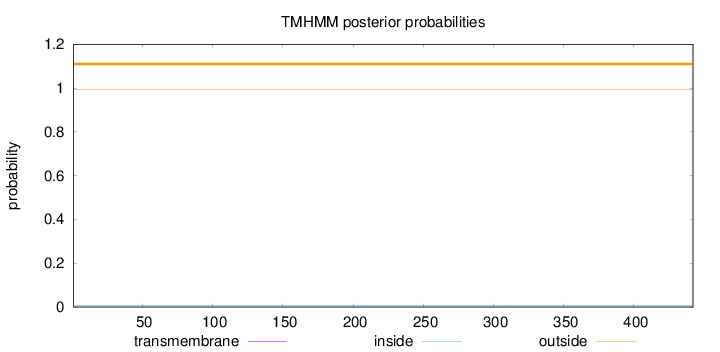
Topology
Length:
442
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00024
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00788
outside
1 - 442
Population Genetic Test Statistics
Pi
366.702807
Theta
193.488933
Tajima's D
3.349165
CLR
0.244193
CSRT
0.991450427478626
Interpretation
Uncertain
