Gene
KWMTBOMO03785
Pre Gene Modal
BGIBMGA010019
Annotation
PREDICTED:_C2_domain-containing_protein_5_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.149
Sequence
CDS
ATGTCCGGGGGCGGCGCGCTGGCGGAGGCGGAGGGCGAGGAGCCGCCCGCGCTGGATCTGTGCGCCGACAAGGACGCTTGCGTGCTGGAACTGGACGAGGCCGAGGACGTGGAGACAGCCAAGTCGCTGTGCGTCCGGCGCGGCCACATGCGCGTGTACAGCCGCCCGCCCCCTGCGCGCGCACCCCCCCTCGCCTTTGCGCAGGTGTGGAGAGCCAGACTGACGTTGTCAGGGCAGGGCGGGGCGGCGTCGCAGGCGGCGGAGAGGCACGTGGCGAGGGCTCTGGACGGAGTCACGTACAAGCTGAGGAGGTTGAACCCCTGCGCGCTGGCTATACCGAAATTCCAACTGGAGTTGCCGGAGGACGAGATCCAGCTGGTGGTGTCTGGCACCGCCATCCCGCTGCCGGAGCCCGGCGCCGCGCCCCGCGACCTGCGCGACGACGACATCTTCGCGCTCGATGAGGAGCAGCTCCTCCGGAACGGACCCGCGCACGACGAGGACAGGAACCTTGTGGGACAGATACCAACGACAGTGTCATTGACAACGCTAAGCGACGTTAGCGGGAGCCGCGCCGTGCGACGGATATGCGCGATCCGGTTACTGTTCGTCAGGGAGACTACGACGGTCCGGGAGATGGGAGGCCTCAGTGGATTTTTACATACGTTCACTTGTGAAGTGCTGGCTATCGCGCGCGCCCACACGGCGGCGCTGGGCGGGAACGCCCTCACGTCGTTCTACCTCACGCAGCTGATGCTACAAGACAACGCCCATAAGAACCAGGGGCAATGTCTTCTATCCGTTGGCGGCGACGTAGTACAGATAACATACTGA
Protein
MSGGGALAEAEGEEPPALDLCADKDACVLELDEAEDVETAKSLCVRRGHMRVYSRPPPARAPPLAFAQVWRARLTLSGQGGAASQAAERHVARALDGVTYKLRRLNPCALAIPKFQLELPEDEIQLVVSGTAIPLPEPGAAPRDLRDDDIFALDEEQLLRNGPAHDEDRNLVGQIPTTVSLTTLSDVSGSRAVRRICAIRLLFVRETTTVREMGGLSGFLHTFTCEVLAIARAHTAALGGNALTSFYLTQLMLQDNAHKNQGQCLLSVGGDVVQITY
Summary
Uniprot
A0A3S2L4U6
A0A2A4IWR1
A0A2A4IWK1
A0A2A4IY55
A0A212EK89
A0A194QEV1
+ More
A0A1A9VDZ6 A0A195EIQ5 A0A0A1WDY2 A0A1B0GF18 U5EQC8 A0A1B0A672 A0A1S4FG53 A0A034VUB5 A0A0A1WTX3 Q172S6 N6T995 A0A0P6IVM0 U4UNJ8 W8BDG6 A0A0K8VV31 A0A1A9XEX4 A0A2J7RPX9 A0A034VTH2 A0A1B0BEV2 A0A151I1I3 W8B2C4 A0A336M205 A0A2J7RPX3 D7EIM0 A0A1I8P0U2 A0A1I8P0S9 W8BDG1 A0A1I8P0N7 A0A1Y1LMR1 A0A1I8P0P5 A0A1W4X3R1 A0A1W4X2K9 E2A3M9 A0A2J7RPX2 A0A1W4WSZ6 A0A067RSU1 A0A1Y1LHT7 A0A026WLH3 A0A3L8D9X2 K7J7A2 A0A195FAQ7 A0A158NKC5 T1PHA2 A0A1I8MRN6 A0A151I8Z2 A0A1Q3G2Q2 A0A1Y1LHS3 A0A1Y1LNM9 A0A1Y1LJW5 A0A1Y1LHT1 A0A1Y1LLK1 A0A1Y1LLI6 A0A1B6EC33 E9IBU0 A0A151WSX8 A0A232EXQ8 A0A2S2NDJ1 A0A2H8TDB0 A0A2S2PR73 A0A2M4AC61 A0A2M4ABR1 A0A2M4AC52 A0A069DWG8 T1IKZ1 A0A0A9VR65 A0A182QW95 A0A0A9VVM2 A0A2M4BBE5 A0A182PH11 A0A0K8SXR4 A0A1E1XGT1 A0A2M4BBC6 A0A2M4BBG3 A0A182XA17 A0A182L9A4 A0A182VIB3 A0A1S4GC29 A0A084VW23 A0A182MTR9 A0A182IXQ2 E2C1M3 A0A2A3ENG1 A0A182N9Z7 A0A023F1S9 A0A182VWC6 A0A1J1HDY5 A0A1J1HFL7 A0A182K2G7 W5JDR7 A0A0V0G8K4 L7MB47
A0A1A9VDZ6 A0A195EIQ5 A0A0A1WDY2 A0A1B0GF18 U5EQC8 A0A1B0A672 A0A1S4FG53 A0A034VUB5 A0A0A1WTX3 Q172S6 N6T995 A0A0P6IVM0 U4UNJ8 W8BDG6 A0A0K8VV31 A0A1A9XEX4 A0A2J7RPX9 A0A034VTH2 A0A1B0BEV2 A0A151I1I3 W8B2C4 A0A336M205 A0A2J7RPX3 D7EIM0 A0A1I8P0U2 A0A1I8P0S9 W8BDG1 A0A1I8P0N7 A0A1Y1LMR1 A0A1I8P0P5 A0A1W4X3R1 A0A1W4X2K9 E2A3M9 A0A2J7RPX2 A0A1W4WSZ6 A0A067RSU1 A0A1Y1LHT7 A0A026WLH3 A0A3L8D9X2 K7J7A2 A0A195FAQ7 A0A158NKC5 T1PHA2 A0A1I8MRN6 A0A151I8Z2 A0A1Q3G2Q2 A0A1Y1LHS3 A0A1Y1LNM9 A0A1Y1LJW5 A0A1Y1LHT1 A0A1Y1LLK1 A0A1Y1LLI6 A0A1B6EC33 E9IBU0 A0A151WSX8 A0A232EXQ8 A0A2S2NDJ1 A0A2H8TDB0 A0A2S2PR73 A0A2M4AC61 A0A2M4ABR1 A0A2M4AC52 A0A069DWG8 T1IKZ1 A0A0A9VR65 A0A182QW95 A0A0A9VVM2 A0A2M4BBE5 A0A182PH11 A0A0K8SXR4 A0A1E1XGT1 A0A2M4BBC6 A0A2M4BBG3 A0A182XA17 A0A182L9A4 A0A182VIB3 A0A1S4GC29 A0A084VW23 A0A182MTR9 A0A182IXQ2 E2C1M3 A0A2A3ENG1 A0A182N9Z7 A0A023F1S9 A0A182VWC6 A0A1J1HDY5 A0A1J1HFL7 A0A182K2G7 W5JDR7 A0A0V0G8K4 L7MB47
Pubmed
EMBL
RSAL01000151
RVE45749.1
NWSH01005605
PCG64099.1
PCG64101.1
PCG64102.1
+ More
AGBW02014315 OWR41890.1 KQ459167 KPJ03475.1 KQ978881 KYN27744.1 GBXI01017083 JAC97208.1 CCAG010012732 GANO01004350 JAB55521.1 GAKP01013834 GAKP01013833 JAC45118.1 GBXI01012196 JAD02096.1 CH477432 EAT41021.1 APGK01039102 KB740967 ENN76819.1 GDUN01000708 JAN95211.1 KB632399 ERL94692.1 GAMC01015374 JAB91181.1 GDHF01009884 GDHF01007793 JAI42430.1 JAI44521.1 NEVH01001354 PNF42883.1 GAKP01013832 JAC45120.1 JXJN01013032 KQ976588 KYM79698.1 GAMC01015377 JAB91178.1 UFQT01000257 SSX22477.1 PNF42881.1 KQ971307 EFA12018.2 GAMC01015379 JAB91176.1 GEZM01055056 GEZM01055052 GEZM01055050 GEZM01055049 GEZM01055048 GEZM01055047 GEZM01055044 JAV73205.1 GL436457 EFN71921.1 PNF42882.1 KK852482 KDR22889.1 GEZM01055046 JAV73209.1 KK107182 EZA55949.1 QOIP01000011 RLU16689.1 KQ981693 KYN37705.1 ADTU01018771 ADTU01018772 ADTU01018773 KA648059 AFP62688.1 KQ978323 KYM95122.1 GFDL01001026 JAV34019.1 GEZM01055051 JAV73199.1 GEZM01055053 JAV73196.1 GEZM01055057 JAV73191.1 GEZM01055055 JAV73194.1 GEZM01055045 JAV73210.1 GEZM01055058 JAV73190.1 GEDC01001800 JAS35498.1 GL762137 EFZ22008.1 KQ982762 KYQ51022.1 NNAY01001717 OXU23143.1 GGMR01002621 MBY15240.1 GFXV01000291 MBW12096.1 GGMR01019353 MBY31972.1 GGFK01004877 MBW38198.1 GGFK01004878 MBW38199.1 GGFK01004867 MBW38188.1 GBGD01000516 JAC88373.1 JH430667 GBHO01045340 JAF98263.1 AXCN02001521 GBHO01045341 GBHO01045339 GDHC01019956 GDHC01017068 JAF98262.1 JAF98264.1 JAP98672.1 JAQ01561.1 GGFJ01001212 MBW50353.1 GBRD01007832 JAG57989.1 GFAC01000947 JAT98241.1 GGFJ01001213 MBW50354.1 GGFJ01001211 MBW50352.1 AAAB01008987 ATLV01017404 ATLV01017405 KE525167 KFB42167.1 AXCM01008204 GL451937 EFN78199.1 KZ288215 PBC32679.1 GBBI01003756 JAC14956.1 CVRI01000001 CRK86205.1 CRK86204.1 ADMH02001846 ETN60919.1 GECL01001610 JAP04514.1 GACK01003789 JAA61245.1
AGBW02014315 OWR41890.1 KQ459167 KPJ03475.1 KQ978881 KYN27744.1 GBXI01017083 JAC97208.1 CCAG010012732 GANO01004350 JAB55521.1 GAKP01013834 GAKP01013833 JAC45118.1 GBXI01012196 JAD02096.1 CH477432 EAT41021.1 APGK01039102 KB740967 ENN76819.1 GDUN01000708 JAN95211.1 KB632399 ERL94692.1 GAMC01015374 JAB91181.1 GDHF01009884 GDHF01007793 JAI42430.1 JAI44521.1 NEVH01001354 PNF42883.1 GAKP01013832 JAC45120.1 JXJN01013032 KQ976588 KYM79698.1 GAMC01015377 JAB91178.1 UFQT01000257 SSX22477.1 PNF42881.1 KQ971307 EFA12018.2 GAMC01015379 JAB91176.1 GEZM01055056 GEZM01055052 GEZM01055050 GEZM01055049 GEZM01055048 GEZM01055047 GEZM01055044 JAV73205.1 GL436457 EFN71921.1 PNF42882.1 KK852482 KDR22889.1 GEZM01055046 JAV73209.1 KK107182 EZA55949.1 QOIP01000011 RLU16689.1 KQ981693 KYN37705.1 ADTU01018771 ADTU01018772 ADTU01018773 KA648059 AFP62688.1 KQ978323 KYM95122.1 GFDL01001026 JAV34019.1 GEZM01055051 JAV73199.1 GEZM01055053 JAV73196.1 GEZM01055057 JAV73191.1 GEZM01055055 JAV73194.1 GEZM01055045 JAV73210.1 GEZM01055058 JAV73190.1 GEDC01001800 JAS35498.1 GL762137 EFZ22008.1 KQ982762 KYQ51022.1 NNAY01001717 OXU23143.1 GGMR01002621 MBY15240.1 GFXV01000291 MBW12096.1 GGMR01019353 MBY31972.1 GGFK01004877 MBW38198.1 GGFK01004878 MBW38199.1 GGFK01004867 MBW38188.1 GBGD01000516 JAC88373.1 JH430667 GBHO01045340 JAF98263.1 AXCN02001521 GBHO01045341 GBHO01045339 GDHC01019956 GDHC01017068 JAF98262.1 JAF98264.1 JAP98672.1 JAQ01561.1 GGFJ01001212 MBW50353.1 GBRD01007832 JAG57989.1 GFAC01000947 JAT98241.1 GGFJ01001213 MBW50354.1 GGFJ01001211 MBW50352.1 AAAB01008987 ATLV01017404 ATLV01017405 KE525167 KFB42167.1 AXCM01008204 GL451937 EFN78199.1 KZ288215 PBC32679.1 GBBI01003756 JAC14956.1 CVRI01000001 CRK86205.1 CRK86204.1 ADMH02001846 ETN60919.1 GECL01001610 JAP04514.1 GACK01003789 JAA61245.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000053268
UP000078200
UP000078492
+ More
UP000092444 UP000092445 UP000008820 UP000019118 UP000030742 UP000092443 UP000235965 UP000092460 UP000078540 UP000007266 UP000095300 UP000192223 UP000000311 UP000027135 UP000053097 UP000279307 UP000002358 UP000078541 UP000005205 UP000095301 UP000078542 UP000075809 UP000215335 UP000075886 UP000075885 UP000076407 UP000075882 UP000075903 UP000030765 UP000075883 UP000075880 UP000008237 UP000242457 UP000075884 UP000075920 UP000183832 UP000075881 UP000000673
UP000092444 UP000092445 UP000008820 UP000019118 UP000030742 UP000092443 UP000235965 UP000092460 UP000078540 UP000007266 UP000095300 UP000192223 UP000000311 UP000027135 UP000053097 UP000279307 UP000002358 UP000078541 UP000005205 UP000095301 UP000078542 UP000075809 UP000215335 UP000075886 UP000075885 UP000076407 UP000075882 UP000075903 UP000030765 UP000075883 UP000075880 UP000008237 UP000242457 UP000075884 UP000075920 UP000183832 UP000075881 UP000000673
PRIDE
Pfam
PF00168 C2
Interpro
SUPFAM
SSF117782
SSF117782
Gene 3D
ProteinModelPortal
A0A3S2L4U6
A0A2A4IWR1
A0A2A4IWK1
A0A2A4IY55
A0A212EK89
A0A194QEV1
+ More
A0A1A9VDZ6 A0A195EIQ5 A0A0A1WDY2 A0A1B0GF18 U5EQC8 A0A1B0A672 A0A1S4FG53 A0A034VUB5 A0A0A1WTX3 Q172S6 N6T995 A0A0P6IVM0 U4UNJ8 W8BDG6 A0A0K8VV31 A0A1A9XEX4 A0A2J7RPX9 A0A034VTH2 A0A1B0BEV2 A0A151I1I3 W8B2C4 A0A336M205 A0A2J7RPX3 D7EIM0 A0A1I8P0U2 A0A1I8P0S9 W8BDG1 A0A1I8P0N7 A0A1Y1LMR1 A0A1I8P0P5 A0A1W4X3R1 A0A1W4X2K9 E2A3M9 A0A2J7RPX2 A0A1W4WSZ6 A0A067RSU1 A0A1Y1LHT7 A0A026WLH3 A0A3L8D9X2 K7J7A2 A0A195FAQ7 A0A158NKC5 T1PHA2 A0A1I8MRN6 A0A151I8Z2 A0A1Q3G2Q2 A0A1Y1LHS3 A0A1Y1LNM9 A0A1Y1LJW5 A0A1Y1LHT1 A0A1Y1LLK1 A0A1Y1LLI6 A0A1B6EC33 E9IBU0 A0A151WSX8 A0A232EXQ8 A0A2S2NDJ1 A0A2H8TDB0 A0A2S2PR73 A0A2M4AC61 A0A2M4ABR1 A0A2M4AC52 A0A069DWG8 T1IKZ1 A0A0A9VR65 A0A182QW95 A0A0A9VVM2 A0A2M4BBE5 A0A182PH11 A0A0K8SXR4 A0A1E1XGT1 A0A2M4BBC6 A0A2M4BBG3 A0A182XA17 A0A182L9A4 A0A182VIB3 A0A1S4GC29 A0A084VW23 A0A182MTR9 A0A182IXQ2 E2C1M3 A0A2A3ENG1 A0A182N9Z7 A0A023F1S9 A0A182VWC6 A0A1J1HDY5 A0A1J1HFL7 A0A182K2G7 W5JDR7 A0A0V0G8K4 L7MB47
A0A1A9VDZ6 A0A195EIQ5 A0A0A1WDY2 A0A1B0GF18 U5EQC8 A0A1B0A672 A0A1S4FG53 A0A034VUB5 A0A0A1WTX3 Q172S6 N6T995 A0A0P6IVM0 U4UNJ8 W8BDG6 A0A0K8VV31 A0A1A9XEX4 A0A2J7RPX9 A0A034VTH2 A0A1B0BEV2 A0A151I1I3 W8B2C4 A0A336M205 A0A2J7RPX3 D7EIM0 A0A1I8P0U2 A0A1I8P0S9 W8BDG1 A0A1I8P0N7 A0A1Y1LMR1 A0A1I8P0P5 A0A1W4X3R1 A0A1W4X2K9 E2A3M9 A0A2J7RPX2 A0A1W4WSZ6 A0A067RSU1 A0A1Y1LHT7 A0A026WLH3 A0A3L8D9X2 K7J7A2 A0A195FAQ7 A0A158NKC5 T1PHA2 A0A1I8MRN6 A0A151I8Z2 A0A1Q3G2Q2 A0A1Y1LHS3 A0A1Y1LNM9 A0A1Y1LJW5 A0A1Y1LHT1 A0A1Y1LLK1 A0A1Y1LLI6 A0A1B6EC33 E9IBU0 A0A151WSX8 A0A232EXQ8 A0A2S2NDJ1 A0A2H8TDB0 A0A2S2PR73 A0A2M4AC61 A0A2M4ABR1 A0A2M4AC52 A0A069DWG8 T1IKZ1 A0A0A9VR65 A0A182QW95 A0A0A9VVM2 A0A2M4BBE5 A0A182PH11 A0A0K8SXR4 A0A1E1XGT1 A0A2M4BBC6 A0A2M4BBG3 A0A182XA17 A0A182L9A4 A0A182VIB3 A0A1S4GC29 A0A084VW23 A0A182MTR9 A0A182IXQ2 E2C1M3 A0A2A3ENG1 A0A182N9Z7 A0A023F1S9 A0A182VWC6 A0A1J1HDY5 A0A1J1HFL7 A0A182K2G7 W5JDR7 A0A0V0G8K4 L7MB47
Ontologies
PANTHER
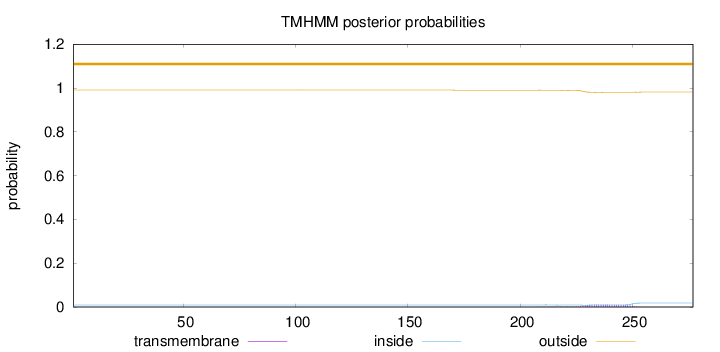
Topology
Length:
277
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26168
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00949
outside
1 - 277
Population Genetic Test Statistics
Pi
378.398302
Theta
172.646084
Tajima's D
3.611681
CLR
0.070589
CSRT
0.995700214989251
Interpretation
Uncertain
