Gene
KWMTBOMO03781
Pre Gene Modal
BGIBMGA010070
Annotation
PREDICTED:_acyl-CoA_synthetase_short-chain_family_member_3?_mitochondrial_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.089
Sequence
CDS
ATGACGTTATTACGAGCGCATTTACAGGTCTCGCGTCTGGCTGGAAAGCTGACAACGTTGGGAGTCAGTAAGGGAGCCCGGGTGCTGATCTACATGCCGCTAGTACCAGAAGCTATTATAGCTATGCTCGCTACCAATCGTATCGGCGCCATTCACTCTGTTGTATTTGGAGGTTTCGCTGCGAGGGAATTAAGAACGAGGATCGAACACGCTGAGCCAACGGTCATCATAGCCGCCAGTTGCGGGGTAGAGCCTAATAAGATCGTCAGGTACAAAGATATTCTGGACGAAGCACTGTGCCAGAGTTCCCATCAGCCTCGCTCCTGCATTATATACCAACGGCGCAGAGTACTAGAGTGTCCCCTTGAAATTGGAAGGGACATATCCTGGGACGAGGGCTTGGAAGCCGACCCGGTACCGTGCGAGTCGGTAGAAGCCAATGAACCCTTGTACATACTATACACATCTGGTGAGTAA
Protein
MTLLRAHLQVSRLAGKLTTLGVSKGARVLIYMPLVPEAIIAMLATNRIGAIHSVVFGGFAARELRTRIEHAEPTVIIAASCGVEPNKIVRYKDILDEALCQSSHQPRSCIIYQRRRVLECPLEIGRDISWDEGLEADPVPCESVEANEPLYILYTSGE
Summary
Uniprot
A0A2H1WB48
A0A194QJ38
A0A2A4JJI7
A0A2H1WLP1
A0A437AVV8
A0A212EK82
+ More
A0A0N1IP08 A0A023F478 E0VNP8 A0A1W4WT96 A0A0V0G4I1 A0A1B6K2Y4 D7EIL8 A0A1B6EB89 A0A1B6LK32 A0A067RG88 A0A1B6GN44 A0A146M909 N6TPX0 U4TXT0 A0A2J7RPX1 A0A182SQR7 A0A2H8TKG3 A0A1Q3G589 A0A182TME7 A0A1Y1K7K9 A0A182L9B4 A0A2J7RPY6 Q7PXM0 A0A182XA15 A0A182HKZ4 A0A182VMV4 W5JDG8 A0A182FRT0 A0A1S3D6F0 A0A232FFW4 A0A2J7RPX8 Q172S4 A0A1S4FG66 J9JL42 A0A2T7PJI5 A0A182K128 K7J7A7 A0A182PH13 A0A182Q2N8 A0A182VWC3 A0A2H8TTP0 A0A182YBA8 A0A182R522 A0A084WNY1 A0A182JFU4 A0A1J1J332 A0A0P5THT2 A0A3R7MAN3 A0A0P5KG23 B0WZJ8 A0A0N8BC78 A0A1B0AIU7 D3TNZ1 A0A1A9VZN2 A0A1B0FND6 A0A1A9V1W4 A0A1B0C444 A0A2C9JWR1 A0A1B0AWT7 A0A1A9Y4W8 A0A352IZW6 A0A1H6QE25 A0A0P5MAK0 E9FTL0 A0A0P5UZ05 A0A164LZA0 A0A0P5EQN5 A0A0P6JIH7 A0A0P5YKU3 A0A0P5VJ86 K9GP14 S6BT24 A0A0P5TEN0 A0A2A3ELU3 A0A3C0JKN5 A0A077MGW1 A0A088A1V7 A0A2T6NG16 A0A182N9Z9 A0A1V4D1A6 A0A060ZL42 A0A349H7F1 A0A1I8NPE0 A0A421K582 A0A1V6GT50 A0A1E2ZD37 K1PDD7 A0A3R7XF06 A0A1B1AW71 A0A1E2UNL2 A0A0H4X786 A0A091ACF5 A0A0P6CCC8
A0A0N1IP08 A0A023F478 E0VNP8 A0A1W4WT96 A0A0V0G4I1 A0A1B6K2Y4 D7EIL8 A0A1B6EB89 A0A1B6LK32 A0A067RG88 A0A1B6GN44 A0A146M909 N6TPX0 U4TXT0 A0A2J7RPX1 A0A182SQR7 A0A2H8TKG3 A0A1Q3G589 A0A182TME7 A0A1Y1K7K9 A0A182L9B4 A0A2J7RPY6 Q7PXM0 A0A182XA15 A0A182HKZ4 A0A182VMV4 W5JDG8 A0A182FRT0 A0A1S3D6F0 A0A232FFW4 A0A2J7RPX8 Q172S4 A0A1S4FG66 J9JL42 A0A2T7PJI5 A0A182K128 K7J7A7 A0A182PH13 A0A182Q2N8 A0A182VWC3 A0A2H8TTP0 A0A182YBA8 A0A182R522 A0A084WNY1 A0A182JFU4 A0A1J1J332 A0A0P5THT2 A0A3R7MAN3 A0A0P5KG23 B0WZJ8 A0A0N8BC78 A0A1B0AIU7 D3TNZ1 A0A1A9VZN2 A0A1B0FND6 A0A1A9V1W4 A0A1B0C444 A0A2C9JWR1 A0A1B0AWT7 A0A1A9Y4W8 A0A352IZW6 A0A1H6QE25 A0A0P5MAK0 E9FTL0 A0A0P5UZ05 A0A164LZA0 A0A0P5EQN5 A0A0P6JIH7 A0A0P5YKU3 A0A0P5VJ86 K9GP14 S6BT24 A0A0P5TEN0 A0A2A3ELU3 A0A3C0JKN5 A0A077MGW1 A0A088A1V7 A0A2T6NG16 A0A182N9Z9 A0A1V4D1A6 A0A060ZL42 A0A349H7F1 A0A1I8NPE0 A0A421K582 A0A1V6GT50 A0A1E2ZD37 K1PDD7 A0A3R7XF06 A0A1B1AW71 A0A1E2UNL2 A0A0H4X786 A0A091ACF5 A0A0P6CCC8
Pubmed
EMBL
ODYU01007475
SOQ50280.1
KQ459167
KPJ03471.1
NWSH01001180
PCG72227.1
+ More
ODYU01009142 SOQ53354.1 RSAL01000347 RVE42302.1 AGBW02014315 OWR41886.1 KQ460869 KPJ11786.1 GBBI01002397 JAC16315.1 DS235341 EEB15004.1 GECL01003536 JAP02588.1 GECU01001894 JAT05813.1 KQ971307 EFA12019.1 GEDC01002107 JAS35191.1 GEBQ01015959 JAT24018.1 KK852482 KDR22891.1 GECZ01005901 JAS63868.1 GDHC01003107 JAQ15522.1 APGK01058936 APGK01058937 APGK01058938 KB741292 ENN70351.1 KB631727 ERL85617.1 NEVH01001354 PNF42887.1 GFXV01002811 MBW14616.1 GFDL01000081 JAV34964.1 GEZM01094463 JAV55675.1 PNF42888.1 AAAB01008987 EAA00842.4 APCN01000876 ADMH02001846 ETN60920.1 NNAY01000294 OXU29410.1 PNF42886.1 CH477432 EAT41023.1 ABLF02019017 PZQS01000003 PVD33552.1 AXCN02001521 GFXV01005147 MBW16952.1 ATLV01024779 KE525360 KFB51925.1 CVRI01000066 CRL05878.1 GDIP01127675 JAL76039.1 QCYY01002281 ROT71552.1 GDIQ01210293 JAK41432.1 DS232209 EDS37512.1 GDIQ01192036 GDIQ01192035 JAK59690.1 EZ423143 ADD19419.1 CCAG010023575 JXJN01025270 JXJN01004952 DNNA01000339 HBC36999.1 FNYH01000001 SEI38477.1 GDIQ01170312 JAK81413.1 GL732524 EFX89382.1 GDIP01106638 JAL97076.1 LRGB01003123 KZS04573.1 GDIP01144732 JAJ78670.1 GDIQ01017730 JAN77007.1 GDIP01056389 JAM47326.1 GDIP01099233 JAM04482.1 ANHY01000030 EKV26469.1 AP012978 BAN69174.1 GDIP01128973 JAL74741.1 KZ288215 PBC32677.1 DMFI01000275 HAI97862.1 CAJC01000202 CCI54872.1 QBVD01000044 PUB83760.1 LAKD02000063 OPF76047.1 LK022848 CDR06820.1 DMWQ01000005 HAS85214.1 NATZ01000024 RLW60586.1 MWEI01000012 OQC28971.1 MARB01000011 ODJ87494.1 JH817576 EKC21892.1 QZAC01000160 RQD81245.1 CP016279 ANP50795.1 LVJZ01000003 ODB96279.1 CP012109 AKQ63727.1 JQIE01000231 KFN37733.1 GDIP01017709 JAM86006.1
ODYU01009142 SOQ53354.1 RSAL01000347 RVE42302.1 AGBW02014315 OWR41886.1 KQ460869 KPJ11786.1 GBBI01002397 JAC16315.1 DS235341 EEB15004.1 GECL01003536 JAP02588.1 GECU01001894 JAT05813.1 KQ971307 EFA12019.1 GEDC01002107 JAS35191.1 GEBQ01015959 JAT24018.1 KK852482 KDR22891.1 GECZ01005901 JAS63868.1 GDHC01003107 JAQ15522.1 APGK01058936 APGK01058937 APGK01058938 KB741292 ENN70351.1 KB631727 ERL85617.1 NEVH01001354 PNF42887.1 GFXV01002811 MBW14616.1 GFDL01000081 JAV34964.1 GEZM01094463 JAV55675.1 PNF42888.1 AAAB01008987 EAA00842.4 APCN01000876 ADMH02001846 ETN60920.1 NNAY01000294 OXU29410.1 PNF42886.1 CH477432 EAT41023.1 ABLF02019017 PZQS01000003 PVD33552.1 AXCN02001521 GFXV01005147 MBW16952.1 ATLV01024779 KE525360 KFB51925.1 CVRI01000066 CRL05878.1 GDIP01127675 JAL76039.1 QCYY01002281 ROT71552.1 GDIQ01210293 JAK41432.1 DS232209 EDS37512.1 GDIQ01192036 GDIQ01192035 JAK59690.1 EZ423143 ADD19419.1 CCAG010023575 JXJN01025270 JXJN01004952 DNNA01000339 HBC36999.1 FNYH01000001 SEI38477.1 GDIQ01170312 JAK81413.1 GL732524 EFX89382.1 GDIP01106638 JAL97076.1 LRGB01003123 KZS04573.1 GDIP01144732 JAJ78670.1 GDIQ01017730 JAN77007.1 GDIP01056389 JAM47326.1 GDIP01099233 JAM04482.1 ANHY01000030 EKV26469.1 AP012978 BAN69174.1 GDIP01128973 JAL74741.1 KZ288215 PBC32677.1 DMFI01000275 HAI97862.1 CAJC01000202 CCI54872.1 QBVD01000044 PUB83760.1 LAKD02000063 OPF76047.1 LK022848 CDR06820.1 DMWQ01000005 HAS85214.1 NATZ01000024 RLW60586.1 MWEI01000012 OQC28971.1 MARB01000011 ODJ87494.1 JH817576 EKC21892.1 QZAC01000160 RQD81245.1 CP016279 ANP50795.1 LVJZ01000003 ODB96279.1 CP012109 AKQ63727.1 JQIE01000231 KFN37733.1 GDIP01017709 JAM86006.1
Proteomes
UP000053268
UP000218220
UP000283053
UP000007151
UP000053240
UP000009046
+ More
UP000192223 UP000007266 UP000027135 UP000019118 UP000030742 UP000235965 UP000075901 UP000075902 UP000075882 UP000007062 UP000076407 UP000075840 UP000075903 UP000000673 UP000069272 UP000079169 UP000215335 UP000008820 UP000007819 UP000245119 UP000075881 UP000002358 UP000075885 UP000075886 UP000075920 UP000076408 UP000075900 UP000030765 UP000075880 UP000183832 UP000283509 UP000002320 UP000092445 UP000091820 UP000092444 UP000078200 UP000092460 UP000076420 UP000092443 UP000242999 UP000000305 UP000076858 UP000009881 UP000015562 UP000242457 UP000035720 UP000005203 UP000244593 UP000075884 UP000033615 UP000095300 UP000284961 UP000094769 UP000005408 UP000284619 UP000092659 UP000094849 UP000009026
UP000192223 UP000007266 UP000027135 UP000019118 UP000030742 UP000235965 UP000075901 UP000075902 UP000075882 UP000007062 UP000076407 UP000075840 UP000075903 UP000000673 UP000069272 UP000079169 UP000215335 UP000008820 UP000007819 UP000245119 UP000075881 UP000002358 UP000075885 UP000075886 UP000075920 UP000076408 UP000075900 UP000030765 UP000075880 UP000183832 UP000283509 UP000002320 UP000092445 UP000091820 UP000092444 UP000078200 UP000092460 UP000076420 UP000092443 UP000242999 UP000000305 UP000076858 UP000009881 UP000015562 UP000242457 UP000035720 UP000005203 UP000244593 UP000075884 UP000033615 UP000095300 UP000284961 UP000094769 UP000005408 UP000284619 UP000092659 UP000094849 UP000009026
Interpro
Gene 3D
ProteinModelPortal
A0A2H1WB48
A0A194QJ38
A0A2A4JJI7
A0A2H1WLP1
A0A437AVV8
A0A212EK82
+ More
A0A0N1IP08 A0A023F478 E0VNP8 A0A1W4WT96 A0A0V0G4I1 A0A1B6K2Y4 D7EIL8 A0A1B6EB89 A0A1B6LK32 A0A067RG88 A0A1B6GN44 A0A146M909 N6TPX0 U4TXT0 A0A2J7RPX1 A0A182SQR7 A0A2H8TKG3 A0A1Q3G589 A0A182TME7 A0A1Y1K7K9 A0A182L9B4 A0A2J7RPY6 Q7PXM0 A0A182XA15 A0A182HKZ4 A0A182VMV4 W5JDG8 A0A182FRT0 A0A1S3D6F0 A0A232FFW4 A0A2J7RPX8 Q172S4 A0A1S4FG66 J9JL42 A0A2T7PJI5 A0A182K128 K7J7A7 A0A182PH13 A0A182Q2N8 A0A182VWC3 A0A2H8TTP0 A0A182YBA8 A0A182R522 A0A084WNY1 A0A182JFU4 A0A1J1J332 A0A0P5THT2 A0A3R7MAN3 A0A0P5KG23 B0WZJ8 A0A0N8BC78 A0A1B0AIU7 D3TNZ1 A0A1A9VZN2 A0A1B0FND6 A0A1A9V1W4 A0A1B0C444 A0A2C9JWR1 A0A1B0AWT7 A0A1A9Y4W8 A0A352IZW6 A0A1H6QE25 A0A0P5MAK0 E9FTL0 A0A0P5UZ05 A0A164LZA0 A0A0P5EQN5 A0A0P6JIH7 A0A0P5YKU3 A0A0P5VJ86 K9GP14 S6BT24 A0A0P5TEN0 A0A2A3ELU3 A0A3C0JKN5 A0A077MGW1 A0A088A1V7 A0A2T6NG16 A0A182N9Z9 A0A1V4D1A6 A0A060ZL42 A0A349H7F1 A0A1I8NPE0 A0A421K582 A0A1V6GT50 A0A1E2ZD37 K1PDD7 A0A3R7XF06 A0A1B1AW71 A0A1E2UNL2 A0A0H4X786 A0A091ACF5 A0A0P6CCC8
A0A0N1IP08 A0A023F478 E0VNP8 A0A1W4WT96 A0A0V0G4I1 A0A1B6K2Y4 D7EIL8 A0A1B6EB89 A0A1B6LK32 A0A067RG88 A0A1B6GN44 A0A146M909 N6TPX0 U4TXT0 A0A2J7RPX1 A0A182SQR7 A0A2H8TKG3 A0A1Q3G589 A0A182TME7 A0A1Y1K7K9 A0A182L9B4 A0A2J7RPY6 Q7PXM0 A0A182XA15 A0A182HKZ4 A0A182VMV4 W5JDG8 A0A182FRT0 A0A1S3D6F0 A0A232FFW4 A0A2J7RPX8 Q172S4 A0A1S4FG66 J9JL42 A0A2T7PJI5 A0A182K128 K7J7A7 A0A182PH13 A0A182Q2N8 A0A182VWC3 A0A2H8TTP0 A0A182YBA8 A0A182R522 A0A084WNY1 A0A182JFU4 A0A1J1J332 A0A0P5THT2 A0A3R7MAN3 A0A0P5KG23 B0WZJ8 A0A0N8BC78 A0A1B0AIU7 D3TNZ1 A0A1A9VZN2 A0A1B0FND6 A0A1A9V1W4 A0A1B0C444 A0A2C9JWR1 A0A1B0AWT7 A0A1A9Y4W8 A0A352IZW6 A0A1H6QE25 A0A0P5MAK0 E9FTL0 A0A0P5UZ05 A0A164LZA0 A0A0P5EQN5 A0A0P6JIH7 A0A0P5YKU3 A0A0P5VJ86 K9GP14 S6BT24 A0A0P5TEN0 A0A2A3ELU3 A0A3C0JKN5 A0A077MGW1 A0A088A1V7 A0A2T6NG16 A0A182N9Z9 A0A1V4D1A6 A0A060ZL42 A0A349H7F1 A0A1I8NPE0 A0A421K582 A0A1V6GT50 A0A1E2ZD37 K1PDD7 A0A3R7XF06 A0A1B1AW71 A0A1E2UNL2 A0A0H4X786 A0A091ACF5 A0A0P6CCC8
PDB
5GXD
E-value=1.3867e-41,
Score=421
Ontologies
GO
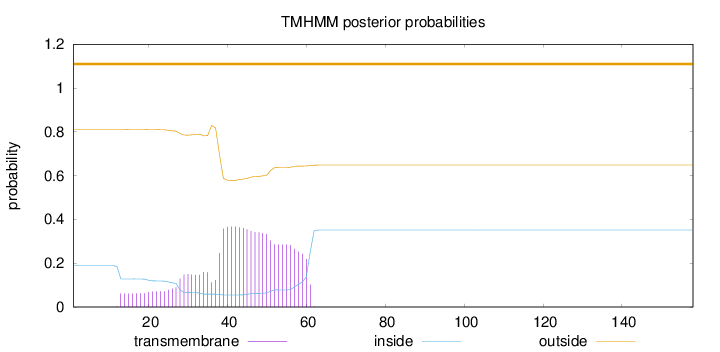
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.7623
Exp number, first 60 AAs:
9.65456
Total prob of N-in:
0.18956
outside
1 - 158
Population Genetic Test Statistics
Pi
276.982873
Theta
166.95449
Tajima's D
2.111156
CLR
0
CSRT
0.897755112244388
Interpretation
Uncertain
