Gene
KWMTBOMO03779 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010067
Annotation
Phosphatidylinositol-binding_clathrin_assembly_protein_LAP_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.568
Sequence
CDS
ATGAACAAGAAGAATTGTCGCGAAGCTCTCGATCTCTATAAAAAGTTCCTGATCCGAATGGATAGAGTTGGGGAGTTCTTGAAGGTAGCGGAGAACGTGGGCATCGACAAGGGCGACATCCCTGACCTCACCAAGGCGCCCAGTAGCCTTCTGGACGCGCTGGAGGGCCACCTCGCCACGCTCGAGGGCAAGAAGGGCTCCGCCGCCAACACTCCCACTCAAACAGCCAGTGCCCAGAAGAACGTGGCCAGCGTCATGGGAGCCCTGTCGTCCACGTCGTCCTCGTTCGGCAACGCGGCCGCCTCCACGCGCCTCGACTACCCGCCCGCCGCCGCCGCGCTGCTCATCGACGACTCGCTGCGACAGCAGGCCCTCGCCGAGGAGGAGGCCGCCATGAACCAGTACAAGGTGCACTTGTTGCAAGGAGACGAGCTGACCGGAAACGAAGACTTGACGGAAATAATCAACAAGGTACAGTCGCCGACGAACGCGGCCAACAACCCGTTCCTAGGCGCGGGGGCTCCGCCCGGCGGCCAGCACGCCATCGTGGACCTGTTCGGGCCCGCGCCCCCCGCCGGCGCCGCCGCCCCGCCAGCCAAGGCCTCCGACGACCTGCTCTCCCTCGGCAACCCCTTCGCCGACATGTTCGGGGCCCCGTCAGCTCCGGCGCAGGGCTCCGCCCCGATGTGGGCGGCGGCGCCGCAGCCCGCGCCCCCCACCAACGGGTTCGCGGCCTTCGCCCCCGCGCCTGCGACACAACAAAACAACACGTTCGTTTCAGAGGCTAACTTCTCTAACGTATTCAATAACACTGACAGCACAGCCGCGGCGCCCAACCCGTTCATGGGCATGGGGGAGTCGTCCCCCTCGCGGCCCCCGCTGCCCCCCCAGCCCGCGCAGCCGCCCAAGTCCGCCTTCGACGAGCTAGAGGACGTGCTCCGCATGTCGCTGGGCTCGCCGGCCAAGTGCCAGCCCGTCACGCAGCAGCCGACGGCGCAGCACCAGTCCGCCGACGTCATGTCGATGCCGATGATGTTCGGCTCGCCGGCCCGCCAGCCGATGGCGCCGATGAGCGGTCACCAGGCGCCGCAGCCCGGCAAGGTGCTCACGGGAGACCTGGACTCGTCCCTCGCGTCGCTCGCTAATAACCTCTCCATCAACAAGACAGCTGCGGCCCAGAAACCGATGCAGTGGAACTCCCCGAAGAACGCGTCGAAGCCGGGACAGACCTGGAGCCCGCAGCCCATGCAGGCCACGACCGGCGCCGGCTACAGGCCGATGGGCCCCGGCATGACCTTACCGACCATGCCCCACACTTTGCCCCACACATACCACCCCCCGCATTATGTTATGCAACAACCCGGCATGGCGATGGGCATGCCCGGAGCCCCCATGATGCCCATGGCGATGGCCGCTCCCATGCGACCCGCCGTACCCCCACAGCCCACCACCAACCAGTTCAGTTAA
Protein
MNKKNCREALDLYKKFLIRMDRVGEFLKVAENVGIDKGDIPDLTKAPSSLLDALEGHLATLEGKKGSAANTPTQTASAQKNVASVMGALSSTSSSFGNAAASTRLDYPPAAAALLIDDSLRQQALAEEEAAMNQYKVHLLQGDELTGNEDLTEIINKVQSPTNAANNPFLGAGAPPGGQHAIVDLFGPAPPAGAAAPPAKASDDLLSLGNPFADMFGAPSAPAQGSAPMWAAAPQPAPPTNGFAAFAPAPATQQNNTFVSEANFSNVFNNTDSTAAAPNPFMGMGESSPSRPPLPPQPAQPPKSAFDELEDVLRMSLGSPAKCQPVTQQPTAQHQSADVMSMPMMFGSPARQPMAPMSGHQAPQPGKVLTGDLDSSLASLANNLSINKTAAAQKPMQWNSPKNASKPGQTWSPQPMQATTGAGYRPMGPGMTLPTMPHTLPHTYHPPHYVMQQPGMAMGMPGAPMMPMAMAAPMRPAVPPQPTTNQFS
Summary
Uniprot
Pubmed
EMBL
BABH01024749
ODYU01001342
SOQ37393.1
JXUM01076339
JXUM01076340
JXUM01076341
+ More
JXUM01076342 JXUM01076343 JXUM01076344 JXUM01076345 KQ562940 KXJ74770.1 AJWK01015500 AJWK01015501 AJWK01015502 AJWK01015503 GANO01000517 JAB59354.1 CM000160 KRK03123.1 AE014297 AGB95721.1 GDHF01029998 GDHF01022119 JAI22316.1 JAI30195.1 GDHF01032289 GDHF01021710 GDHF01011554 GDHF01008958 GDHF01002767 GDHF01002669 JAI20025.1 JAI30604.1 JAI40760.1 JAI43356.1 JAI49547.1 JAI49645.1 GFDF01006392 JAV07692.1 GEDC01025917 JAS11381.1 AJVK01006463 AJVK01006464 AJVK01006465 AJVK01006466 GL447829 EFN85867.1
JXUM01076342 JXUM01076343 JXUM01076344 JXUM01076345 KQ562940 KXJ74770.1 AJWK01015500 AJWK01015501 AJWK01015502 AJWK01015503 GANO01000517 JAB59354.1 CM000160 KRK03123.1 AE014297 AGB95721.1 GDHF01029998 GDHF01022119 JAI22316.1 JAI30195.1 GDHF01032289 GDHF01021710 GDHF01011554 GDHF01008958 GDHF01002767 GDHF01002669 JAI20025.1 JAI30604.1 JAI40760.1 JAI43356.1 JAI49547.1 JAI49645.1 GFDF01006392 JAV07692.1 GEDC01025917 JAS11381.1 AJVK01006463 AJVK01006464 AJVK01006465 AJVK01006466 GL447829 EFN85867.1
Proteomes
PRIDE
Pfam
PF07651 ANTH
SUPFAM
SSF48464
SSF48464
Gene 3D
ProteinModelPortal
PDB
1HX8
E-value=5.1233e-26,
Score=293
Ontologies
GO
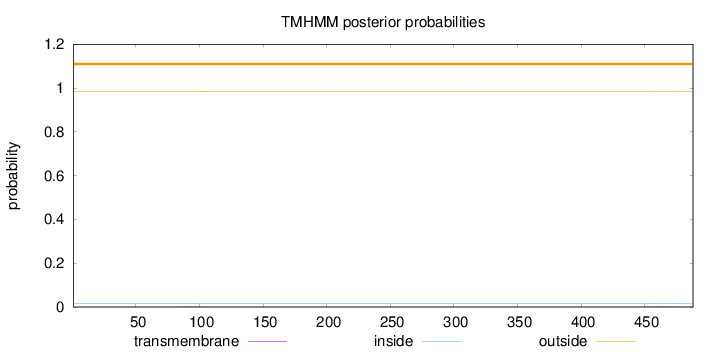
Topology
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0697900000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01701
outside
1 - 488
Population Genetic Test Statistics
Pi
287.409489
Theta
192.795975
Tajima's D
1.407332
CLR
0.007584
CSRT
0.769211539423029
Interpretation
Uncertain
