Gene
KWMTBOMO03775
Pre Gene Modal
BGIBMGA002957
Annotation
PREDICTED:_zinc_finger_CCHC_domain-containing_protein_13-like_[Bombyx_mori]
Full name
Gag polyprotein
Location in the cell
Nuclear Reliability : 2.018
Sequence
CDS
ATGCGACGTGTCACCCACACCGATGCGGAGGTTATTCCAACGTTCCGGCCGGATGAAAAATCTAGCAACGTCAAGGGATGGCTGCACAAGATCGACCAGTTGGATCACGTATATGGATGGGACAACAAAGACTGCCAGTTCATCATGCAGATATGTCTTCGTGGGTCGGCTAGGGATTGTGCGATACACCACCGCAACTATACTACACATCCGCGAATGGCTACCACATGGAGAAGAGAAGGTGCTGTTGCTCCAGTTGCAAGAGGGACCGACTTTCAACCAAAGAAATGTTACGCCTGCCGAAGAGAAGGTCATGAAACAAAGAACTGCAAAGAGCCGCGCTGCGAGGTGTGCCATCGCCCGGGACACACGTCGGTCAGCTGA
Protein
MRRVTHTDAEVIPTFRPDEKSSNVKGWLHKIDQLDHVYGWDNKDCQFIMQICLRGSARDCAIHHRNYTTHPRMATTWRREGAVAPVARGTDFQPKKCYACRREGHETKNCKEPRCEVCHRPGHTSVS
Summary
Catalytic Activity
3'-end directed exonucleolytic cleavage of viral RNA-DNA hybrid.
Endohydrolysis of RNA in RNA/DNA hybrids. Three different cleavage modes: 1. sequence-specific internal cleavage of RNA. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction. 2. RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end. 3. DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Endohydrolysis of RNA in RNA/DNA hybrids. Three different cleavage modes: 1. sequence-specific internal cleavage of RNA. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction. 2. RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end. 3. DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Cofactor
Mg(2+)
Similarity
Belongs to the primate lentivirus group gag polyprotein family.
Uniprot
H9J0C1
H9JKH1
V9QIN8
Q87621
Q79351
Q8ADE9
+ More
Q8ADV1 C9DWL7 A0A386QZH4 Q7SPP5 Q8AC28 Q80634 D3X833 A9Y691 C9DX50 Q8AC47 Q9J6W5 U6BH18 B6DQT6 Q7ZNK7 A0A1I9WBM0 C7G2K5 Q5U6W1 Q6JFX9 D3X851 A0A386QZJ2 Q5U6V8 W5VTT7 A0A2L1KWD2 D8X027 A0A386QZS4 A0A0U4DFU7 C9DX69 A0A1B3YUV4 W5VSJ1 C9DX63 C9DWB8 Q5Q9E1 A0A2L1KXV4 Q8AC39 A0A3S7RKE9 A0A3S7RK58 C7ASC4 A0A386QZI7 A0A1B3YUZ5 A0A1B3YV34 A0A1B3YUZ9
Q8ADV1 C9DWL7 A0A386QZH4 Q7SPP5 Q8AC28 Q80634 D3X833 A9Y691 C9DX50 Q8AC47 Q9J6W5 U6BH18 B6DQT6 Q7ZNK7 A0A1I9WBM0 C7G2K5 Q5U6W1 Q6JFX9 D3X851 A0A386QZJ2 Q5U6V8 W5VTT7 A0A2L1KWD2 D8X027 A0A386QZS4 A0A0U4DFU7 C9DX69 A0A1B3YUV4 W5VSJ1 C9DX63 C9DWB8 Q5Q9E1 A0A2L1KXV4 Q8AC39 A0A3S7RKE9 A0A3S7RK58 C7ASC4 A0A386QZI7 A0A1B3YUZ5 A0A1B3YV34 A0A1B3YUZ9
Pubmed
EMBL
BABH01010590
BABH01024735
BABH01024736
KF859747
AHC30187.1
U42720
+ More
AAB47723.1 L11769 AAA44689.1 AF484513 AAN73754.1 AF484494 AAN73583.1 GQ431214 ACV94279.1 MH971628 AYE55483.1 AJ519489 CAD59562.1 AAN73755.1 L11796 AAA45298.1 GU245717 ADD31665.1 EU110090 ABW86715.1 GQ431487 ACV94462.1 AAN73584.1 AF184496 AAF28628.1 KF716490 AHA33898.1 FJ155131 ACI05374.1 AJ488927 CAD44561.1 CAD58643.1 KU921966 APA29369.1 AB485658 AB485659 BAH97586.1 AY803367 AAV50140.1 AY492788 AAT06727.1 GU245738 ADD31683.1 MH971621 AYE55476.1 AY803372 AAV50143.1 KF823025 AHH85792.1 MF109468 AVE26833.1 GU458744 ADJ58364.1 MH971715 AYE55570.1 KU168277 ALX35278.1 GQ431509 ACV94481.1 KX302424 AOH69338.1 KF823032 AHH85799.1 GQ431503 ACV94475.1 GQ430961 ACV94180.1 AY772965 AAV85025.1 MF109656 AVE27377.1 AF484502 AAN73656.1 MG989663 AXP13981.1 MG989641 AXP13876.1 FJ606395 ACU31735.1 MH971638 AYE55493.1 KX302467 AOH69374.1 KX302509 AOH69406.1 KX302466 AOH69372.1
AAB47723.1 L11769 AAA44689.1 AF484513 AAN73754.1 AF484494 AAN73583.1 GQ431214 ACV94279.1 MH971628 AYE55483.1 AJ519489 CAD59562.1 AAN73755.1 L11796 AAA45298.1 GU245717 ADD31665.1 EU110090 ABW86715.1 GQ431487 ACV94462.1 AAN73584.1 AF184496 AAF28628.1 KF716490 AHA33898.1 FJ155131 ACI05374.1 AJ488927 CAD44561.1 CAD58643.1 KU921966 APA29369.1 AB485658 AB485659 BAH97586.1 AY803367 AAV50140.1 AY492788 AAT06727.1 GU245738 ADD31683.1 MH971621 AYE55476.1 AY803372 AAV50143.1 KF823025 AHH85792.1 MF109468 AVE26833.1 GU458744 ADJ58364.1 MH971715 AYE55570.1 KU168277 ALX35278.1 GQ431509 ACV94481.1 KX302424 AOH69338.1 KF823032 AHH85799.1 GQ431503 ACV94475.1 GQ430961 ACV94180.1 AY772965 AAV85025.1 MF109656 AVE27377.1 AF484502 AAN73656.1 MG989663 AXP13981.1 MG989641 AXP13876.1 FJ606395 ACU31735.1 MH971638 AYE55493.1 KX302467 AOH69374.1 KX302509 AOH69406.1 KX302466 AOH69372.1
Proteomes
Pfam
Interpro
IPR036875
Znf_CCHC_sf
+ More
IPR001878 Znf_CCHC
IPR014817 Gag_p6
IPR000071 Lentvrl_matrix_N
IPR010999 Retrovr_matrix
IPR012344 Matrix_HIV/RSV_N
IPR000721 Gag_p24
IPR008916 Retrov_capsid_C
IPR008919 Retrov_capsid_N
IPR010659 RVT_connect
IPR012337 RNaseH-like_sf
IPR036862 Integrase_C_dom_sf_retrovir
IPR001969 Aspartic_peptidase_AS
IPR001584 Integrase_cat-core
IPR003308 Integrase_Zn-bd_dom_N
IPR001995 Peptidase_A2_cat
IPR000477 RT_dom
IPR010661 RVT_thumb
IPR002156 RNaseH_domain
IPR018061 Retropepsins
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001037 Integrase_C_retrovir
IPR034170 Retropepsin-like_cat_dom
IPR017856 Integrase-like_N
IPR001878 Znf_CCHC
IPR014817 Gag_p6
IPR000071 Lentvrl_matrix_N
IPR010999 Retrovr_matrix
IPR012344 Matrix_HIV/RSV_N
IPR000721 Gag_p24
IPR008916 Retrov_capsid_C
IPR008919 Retrov_capsid_N
IPR010659 RVT_connect
IPR012337 RNaseH-like_sf
IPR036862 Integrase_C_dom_sf_retrovir
IPR001969 Aspartic_peptidase_AS
IPR001584 Integrase_cat-core
IPR003308 Integrase_Zn-bd_dom_N
IPR001995 Peptidase_A2_cat
IPR000477 RT_dom
IPR010661 RVT_thumb
IPR002156 RNaseH_domain
IPR018061 Retropepsins
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR001037 Integrase_C_retrovir
IPR034170 Retropepsin-like_cat_dom
IPR017856 Integrase-like_N
SUPFAM
ProteinModelPortal
H9J0C1
H9JKH1
V9QIN8
Q87621
Q79351
Q8ADE9
+ More
Q8ADV1 C9DWL7 A0A386QZH4 Q7SPP5 Q8AC28 Q80634 D3X833 A9Y691 C9DX50 Q8AC47 Q9J6W5 U6BH18 B6DQT6 Q7ZNK7 A0A1I9WBM0 C7G2K5 Q5U6W1 Q6JFX9 D3X851 A0A386QZJ2 Q5U6V8 W5VTT7 A0A2L1KWD2 D8X027 A0A386QZS4 A0A0U4DFU7 C9DX69 A0A1B3YUV4 W5VSJ1 C9DX63 C9DWB8 Q5Q9E1 A0A2L1KXV4 Q8AC39 A0A3S7RKE9 A0A3S7RK58 C7ASC4 A0A386QZI7 A0A1B3YUZ5 A0A1B3YV34 A0A1B3YUZ9
Q8ADV1 C9DWL7 A0A386QZH4 Q7SPP5 Q8AC28 Q80634 D3X833 A9Y691 C9DX50 Q8AC47 Q9J6W5 U6BH18 B6DQT6 Q7ZNK7 A0A1I9WBM0 C7G2K5 Q5U6W1 Q6JFX9 D3X851 A0A386QZJ2 Q5U6V8 W5VTT7 A0A2L1KWD2 D8X027 A0A386QZS4 A0A0U4DFU7 C9DX69 A0A1B3YUV4 W5VSJ1 C9DX63 C9DWB8 Q5Q9E1 A0A2L1KXV4 Q8AC39 A0A3S7RKE9 A0A3S7RK58 C7ASC4 A0A386QZI7 A0A1B3YUZ5 A0A1B3YV34 A0A1B3YUZ9
PDB
5O2U
E-value=0.0120424,
Score=84
Ontologies
GO
GO:0008270
GO:0003676
GO:0005198
GO:0042025
GO:0019013
GO:0030430
GO:0039702
GO:0003723
GO:0016032
GO:0004533
GO:0039657
GO:0046718
GO:0004190
GO:0006310
GO:0003964
GO:0016020
GO:0075732
GO:0020002
GO:0003677
GO:0015074
GO:0003887
GO:0044826
GO:0075713
GO:0004523
GO:0006811
GO:0007155
GO:0000049
GO:0004812
GO:0043039
GO:0042555
GO:0048384
Topology
Subcellular location
Virion
Host nucleus
Host cytoplasm
Host cell membrane
Host nucleus
Host cytoplasm
Host cell membrane
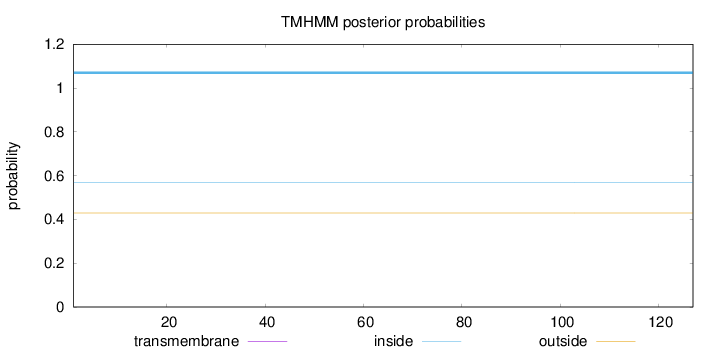
Length:
127
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.57018
inside
1 - 127
Population Genetic Test Statistics
Pi
462.410965
Theta
228.715638
Tajima's D
3.209528
CLR
0
CSRT
0.986750662466877
Interpretation
Uncertain
