Gene
KWMTBOMO03770
Pre Gene Modal
BGIBMGA010063
Annotation
PREDICTED:_trypsin-like_serine_protease_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 3.52
Sequence
CDS
ATGGCTTACGTCGCTTTGGTGCTCTTGTTGGTGGCAGCCGTGAGCTGCTCCAGCAATGATATCATTGGCACTCCCGTTGAAGTCACAACGTTTCCCAGTATTGTCCAGGTTGAGATCCGCGTAGGTGATTATTGGATCCAGAAGTGCGCTGGAGGTATCATCACCAATTACCACATCGTCTCTGTCGCCACGTGTTTTAGTGGAGCTAACTACAGTCCAGAACGCCGTCGAGTTAGAGCAGGCGCCAGTCGACGCGGTGAAGCCGGAAGTATTGTCTACGTTATGCAGGAATACAACCACTATTACTTCGGTTCCTTGGACGCTGACGGTGACATAAGCGTCGTCCGTCTCACCGGTCCCCTGACCTTCAGTACCGGCATAGCTCAGGCTCCTATTGTCGGACAGGGGATTGAGCTGCCTTCAGGACTTAAAGTTGTGAACGTTGGCTGGGGCACCACTGTTCAAGGGAGCCAGGACGTCGACGACGCCTTGTACAAACTCGACCTATTTACTCAGAATAATGACCGCTGTGTCGTTAAGTACCTTTATGCAACTAACCCCAGAGTCGTTACCGAGAATATGATCTGTGCCGGTCTGTACGGGAAAGGTCGGGACTTTGACGTCAGTGACCTTGGTTCCCCCCTCTACTATGACAACGTCCTAGTGGGTCTCCTCTCGTTTGGCGTGCCTAACGCTGATGATGAGTACCCTGTTGTCGCTACTGCTGTATCGTTTTTCAGTAACTGGATCACCAGAACCGCCGTTTAA
Protein
MAYVALVLLLVAAVSCSSNDIIGTPVEVTTFPSIVQVEIRVGDYWIQKCAGGIITNYHIVSVATCFSGANYSPERRRVRAGASRRGEAGSIVYVMQEYNHYYFGSLDADGDISVVRLTGPLTFSTGIAQAPIVGQGIELPSGLKVVNVGWGTTVQGSQDVDDALYKLDLFTQNNDRCVVKYLYATNPRVVTENMICAGLYGKGRDFDVSDLGSPLYYDNVLVGLLSFGVPNADDEYPVVATAVSFFSNWITRTAV
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JKL2
A0A2H1W3R1
A0A2H1VGN5
A0A2W1BN83
A0A2A4JTK1
C9W8I5
+ More
G8GBC4 Q1HPT9 A0A0L7L5I1 H9JKH3 H9JKL1 I7DEJ5 A0A2A4JUT0 A0A2W1BJP2 A0A1B1LTR2 A0A194QET8 A0A3S2N5V1 A0A2W1BTM0 A0A2H1VGP3 B4X9A2 A0A2A4JTX8 C9W8I6 A0A194QD40 A0A212EW56 A0A2A4JTP3 A0A2H1W3L1 A0A2H1WEE4 Q56IB1 B1NLD8 C9W8J1 Q56IB2 B6CME9 A0A194QJ28 A0A194QDG3 Q56IB0 A0A0L7L171 A0A2H1VGQ2 A0A1B0RHN8 B6D1Q8 A0A194QDC5 A0A2W1BP62 Q9U4I4 A0A2W1BN80 B1NLE4 A0A0L7KYH0 A0A437BSH3 O18439 A0A437BTC7 A0A2A4JSY0 A9P3S4 A5CG75 A0A089RSX5 A0A0L7L163 I7C7Q2 Q9U4I5 A0A0N1I8C7 E0W9N5 A0A2H1VIA8 I6TEW4 Q9NB81 Q4L1L0 A0A0M4M544 B6D1Q7 A0A223AIS8 B4X9A4 B4X9A3 A0A2A4JUR5 A0A0M4MT18 Q6R560 A0A2H1VGQ8 A0A2H1W591 Q4L1L3 A0A1E1W498 Q9NB80 Q4L1L1 A0A1E1WB94 A0A194QD45 A0A0N1PGP3 Q9NB89 A0A1E1WML3 E2IGY7 H9JKL0 Q4L1L4 A0A2U8NFH1 Q9NB87 A0A2H1VGM7 Q4L1L5 A0A2W1BSQ0 Q4L1K9 O45047 A0A2A4JCA8 A0A2H1VG89 A0A2W1BP46 I7D523
G8GBC4 Q1HPT9 A0A0L7L5I1 H9JKH3 H9JKL1 I7DEJ5 A0A2A4JUT0 A0A2W1BJP2 A0A1B1LTR2 A0A194QET8 A0A3S2N5V1 A0A2W1BTM0 A0A2H1VGP3 B4X9A2 A0A2A4JTX8 C9W8I6 A0A194QD40 A0A212EW56 A0A2A4JTP3 A0A2H1W3L1 A0A2H1WEE4 Q56IB1 B1NLD8 C9W8J1 Q56IB2 B6CME9 A0A194QJ28 A0A194QDG3 Q56IB0 A0A0L7L171 A0A2H1VGQ2 A0A1B0RHN8 B6D1Q8 A0A194QDC5 A0A2W1BP62 Q9U4I4 A0A2W1BN80 B1NLE4 A0A0L7KYH0 A0A437BSH3 O18439 A0A437BTC7 A0A2A4JSY0 A9P3S4 A5CG75 A0A089RSX5 A0A0L7L163 I7C7Q2 Q9U4I5 A0A0N1I8C7 E0W9N5 A0A2H1VIA8 I6TEW4 Q9NB81 Q4L1L0 A0A0M4M544 B6D1Q7 A0A223AIS8 B4X9A4 B4X9A3 A0A2A4JUR5 A0A0M4MT18 Q6R560 A0A2H1VGQ8 A0A2H1W591 Q4L1L3 A0A1E1W498 Q9NB80 Q4L1L1 A0A1E1WB94 A0A194QD45 A0A0N1PGP3 Q9NB89 A0A1E1WML3 E2IGY7 H9JKL0 Q4L1L4 A0A2U8NFH1 Q9NB87 A0A2H1VGM7 Q4L1L5 A0A2W1BSQ0 Q4L1K9 O45047 A0A2A4JCA8 A0A2H1VG89 A0A2W1BP46 I7D523
Pubmed
EMBL
BABH01024728
BABH01024729
ODYU01006115
SOQ47673.1
ODYU01002467
SOQ39995.1
+ More
KZ150010 PZC75085.1 NWSH01000631 PCG75149.1 FJ205431 ACR15998.2 BABH01024734 JN120256 AER70119.1 DQ443313 KM027249 ABF51402.1 AIU94618.1 JTDY01002795 KOB70712.1 BABH01024730 BABH01024727 JN793549 AFO68329.1 PCG75152.1 PZC75088.1 KX380991 ANS56507.1 KQ459167 KPJ03460.1 RSAL01000347 RVE42309.1 PZC75083.1 SOQ39998.1 EF531638 ABR88249.1 PCG75148.1 FJ205433 ACR15999.2 KPJ03458.1 AGBW02012089 OWR45697.1 PCG75146.1 SOQ47675.1 ODYU01007983 SOQ51222.1 AY953066 AAX62039.1 EF600053 ABU98618.1 PZC75086.1 FJ205440 ACR16004.1 AY953065 AAX62038.1 EU325549 ACB54939.1 KPJ03461.1 KPJ03459.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JTDY01003642 KOB69232.1 SOQ39997.1 KM360185 AKH49601.1 EU673451 ACI45414.1 KPJ03462.1 PZC75087.1 AF173498 AAF24228.1 PZC75084.1 EF600059 ABU98624.1 JTDY01004416 KOB68185.1 RSAL01000013 RVE53438.1 Y12274 CAA72953.1 RVE53439.1 PCG75147.1 EF104913 ABP96915.1 AM690450 CAM84320.1 KM083776 AIR09757.1 KOB69233.1 JN793548 AFO68328.1 AF173497 AAF24227.1 KQ460409 KPJ14944.1 FJ205419 ADM35106.1 SOQ39994.1 JQ904129 AFM77759.1 AF261981 AAF74743.1 AY587155 AAT95347.1 KR024672 ALE15214.1 EU673450 ACI45413.1 KX951432 KY684226 ARR75600.1 ASS33865.1 EF531640 ABR88251.1 EF531639 EF531641 ABR88250.1 ABR88252.1 PCG75150.1 KR024671 ALE15213.1 AY513650 AAR98919.1 SOQ39996.1 SOQ47674.1 AY587152 AAT95344.1 GDQN01009270 JAT81784.1 AF261982 AAF74744.1 AY587154 AAT95346.1 GDQN01006776 JAT84278.1 KPJ03463.1 KPJ14943.1 AF261973 AAF74735.1 GDQN01011173 GDQN01002795 JAT79881.1 JAT88259.1 HM535626 ADK66277.1 BABH01024725 AY587151 AAT95343.1 MF407319 AWL83215.1 AF261975 AAF74737.1 SOQ39999.1 AY587150 AAT95342.1 KZ149911 PZC78102.1 AY587156 AAT95348.1 AF045142 AAC02220.1 NWSH01002022 PCG69396.1 ODYU01002419 SOQ39873.1 KZ150104 PZC73453.1 JN793545 AFO68325.1
KZ150010 PZC75085.1 NWSH01000631 PCG75149.1 FJ205431 ACR15998.2 BABH01024734 JN120256 AER70119.1 DQ443313 KM027249 ABF51402.1 AIU94618.1 JTDY01002795 KOB70712.1 BABH01024730 BABH01024727 JN793549 AFO68329.1 PCG75152.1 PZC75088.1 KX380991 ANS56507.1 KQ459167 KPJ03460.1 RSAL01000347 RVE42309.1 PZC75083.1 SOQ39998.1 EF531638 ABR88249.1 PCG75148.1 FJ205433 ACR15999.2 KPJ03458.1 AGBW02012089 OWR45697.1 PCG75146.1 SOQ47675.1 ODYU01007983 SOQ51222.1 AY953066 AAX62039.1 EF600053 ABU98618.1 PZC75086.1 FJ205440 ACR16004.1 AY953065 AAX62038.1 EU325549 ACB54939.1 KPJ03461.1 KPJ03459.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JTDY01003642 KOB69232.1 SOQ39997.1 KM360185 AKH49601.1 EU673451 ACI45414.1 KPJ03462.1 PZC75087.1 AF173498 AAF24228.1 PZC75084.1 EF600059 ABU98624.1 JTDY01004416 KOB68185.1 RSAL01000013 RVE53438.1 Y12274 CAA72953.1 RVE53439.1 PCG75147.1 EF104913 ABP96915.1 AM690450 CAM84320.1 KM083776 AIR09757.1 KOB69233.1 JN793548 AFO68328.1 AF173497 AAF24227.1 KQ460409 KPJ14944.1 FJ205419 ADM35106.1 SOQ39994.1 JQ904129 AFM77759.1 AF261981 AAF74743.1 AY587155 AAT95347.1 KR024672 ALE15214.1 EU673450 ACI45413.1 KX951432 KY684226 ARR75600.1 ASS33865.1 EF531640 ABR88251.1 EF531639 EF531641 ABR88250.1 ABR88252.1 PCG75150.1 KR024671 ALE15213.1 AY513650 AAR98919.1 SOQ39996.1 SOQ47674.1 AY587152 AAT95344.1 GDQN01009270 JAT81784.1 AF261982 AAF74744.1 AY587154 AAT95346.1 GDQN01006776 JAT84278.1 KPJ03463.1 KPJ14943.1 AF261973 AAF74735.1 GDQN01011173 GDQN01002795 JAT79881.1 JAT88259.1 HM535626 ADK66277.1 BABH01024725 AY587151 AAT95343.1 MF407319 AWL83215.1 AF261975 AAF74737.1 SOQ39999.1 AY587150 AAT95342.1 KZ149911 PZC78102.1 AY587156 AAT95348.1 AF045142 AAC02220.1 NWSH01002022 PCG69396.1 ODYU01002419 SOQ39873.1 KZ150104 PZC73453.1 JN793545 AFO68325.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JKL2
A0A2H1W3R1
A0A2H1VGN5
A0A2W1BN83
A0A2A4JTK1
C9W8I5
+ More
G8GBC4 Q1HPT9 A0A0L7L5I1 H9JKH3 H9JKL1 I7DEJ5 A0A2A4JUT0 A0A2W1BJP2 A0A1B1LTR2 A0A194QET8 A0A3S2N5V1 A0A2W1BTM0 A0A2H1VGP3 B4X9A2 A0A2A4JTX8 C9W8I6 A0A194QD40 A0A212EW56 A0A2A4JTP3 A0A2H1W3L1 A0A2H1WEE4 Q56IB1 B1NLD8 C9W8J1 Q56IB2 B6CME9 A0A194QJ28 A0A194QDG3 Q56IB0 A0A0L7L171 A0A2H1VGQ2 A0A1B0RHN8 B6D1Q8 A0A194QDC5 A0A2W1BP62 Q9U4I4 A0A2W1BN80 B1NLE4 A0A0L7KYH0 A0A437BSH3 O18439 A0A437BTC7 A0A2A4JSY0 A9P3S4 A5CG75 A0A089RSX5 A0A0L7L163 I7C7Q2 Q9U4I5 A0A0N1I8C7 E0W9N5 A0A2H1VIA8 I6TEW4 Q9NB81 Q4L1L0 A0A0M4M544 B6D1Q7 A0A223AIS8 B4X9A4 B4X9A3 A0A2A4JUR5 A0A0M4MT18 Q6R560 A0A2H1VGQ8 A0A2H1W591 Q4L1L3 A0A1E1W498 Q9NB80 Q4L1L1 A0A1E1WB94 A0A194QD45 A0A0N1PGP3 Q9NB89 A0A1E1WML3 E2IGY7 H9JKL0 Q4L1L4 A0A2U8NFH1 Q9NB87 A0A2H1VGM7 Q4L1L5 A0A2W1BSQ0 Q4L1K9 O45047 A0A2A4JCA8 A0A2H1VG89 A0A2W1BP46 I7D523
G8GBC4 Q1HPT9 A0A0L7L5I1 H9JKH3 H9JKL1 I7DEJ5 A0A2A4JUT0 A0A2W1BJP2 A0A1B1LTR2 A0A194QET8 A0A3S2N5V1 A0A2W1BTM0 A0A2H1VGP3 B4X9A2 A0A2A4JTX8 C9W8I6 A0A194QD40 A0A212EW56 A0A2A4JTP3 A0A2H1W3L1 A0A2H1WEE4 Q56IB1 B1NLD8 C9W8J1 Q56IB2 B6CME9 A0A194QJ28 A0A194QDG3 Q56IB0 A0A0L7L171 A0A2H1VGQ2 A0A1B0RHN8 B6D1Q8 A0A194QDC5 A0A2W1BP62 Q9U4I4 A0A2W1BN80 B1NLE4 A0A0L7KYH0 A0A437BSH3 O18439 A0A437BTC7 A0A2A4JSY0 A9P3S4 A5CG75 A0A089RSX5 A0A0L7L163 I7C7Q2 Q9U4I5 A0A0N1I8C7 E0W9N5 A0A2H1VIA8 I6TEW4 Q9NB81 Q4L1L0 A0A0M4M544 B6D1Q7 A0A223AIS8 B4X9A4 B4X9A3 A0A2A4JUR5 A0A0M4MT18 Q6R560 A0A2H1VGQ8 A0A2H1W591 Q4L1L3 A0A1E1W498 Q9NB80 Q4L1L1 A0A1E1WB94 A0A194QD45 A0A0N1PGP3 Q9NB89 A0A1E1WML3 E2IGY7 H9JKL0 Q4L1L4 A0A2U8NFH1 Q9NB87 A0A2H1VGM7 Q4L1L5 A0A2W1BSQ0 Q4L1K9 O45047 A0A2A4JCA8 A0A2H1VG89 A0A2W1BP46 I7D523
PDB
1FXY
E-value=1.86063e-12,
Score=173
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.997534
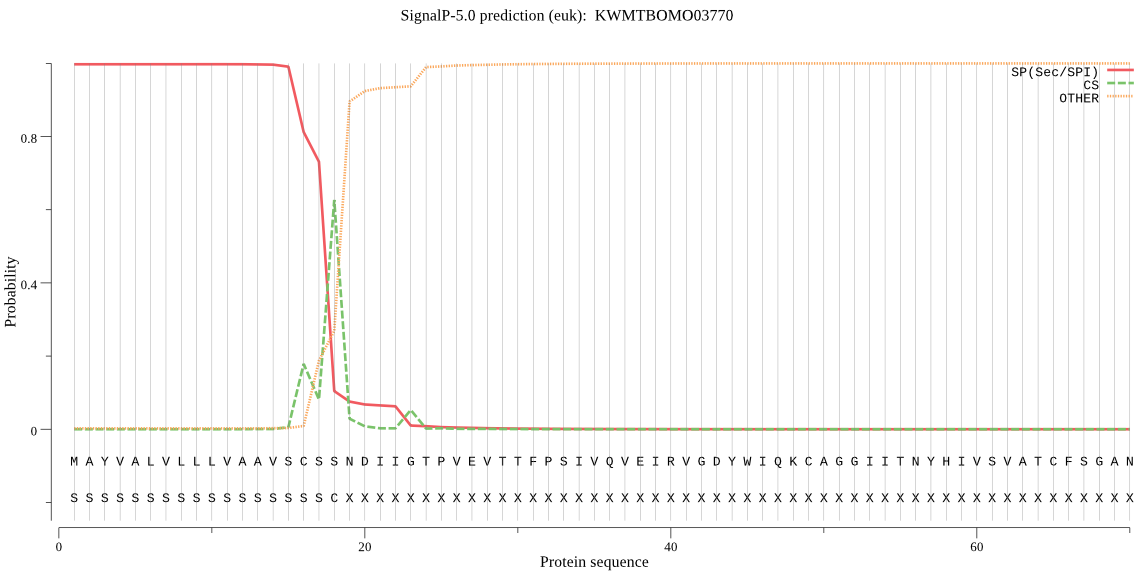
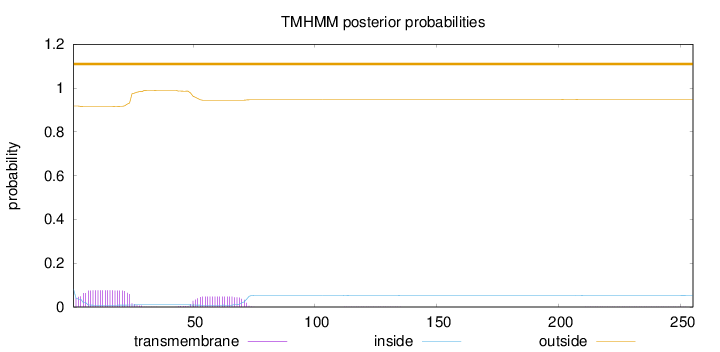
Length:
255
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.70082000000001
Exp number, first 60 AAs:
2.15741
Total prob of N-in:
0.08218
outside
1 - 255
Population Genetic Test Statistics
Pi
409.978084
Theta
198.748488
Tajima's D
-1.626124
CLR
0.659362
CSRT
0.0433478326083696
Interpretation
Uncertain
