Gene
KWMTBOMO03769 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010062
Annotation
trypsin-like_protease_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 4.198
Sequence
CDS
ATGGTTTCCGTTCTCACCTTAGCCGTAGCCGTGCTCGCTGCCGTCTGCAGCTGCACTGCTAGCAATCTCGGTAGACCTGTGTCTATTGGCGAGCACCCCAGCCTGGTCCAAATTGAAGTATTCCTACCCATCTTGAATCAATGGTTCCAACAGTGCGCTGGTATTGTTCTCACCAACTACCACTACCTTTCAACTGCTACCTGTTTCCATGGAGAATTCTACGATCCTGCATACCGTCGCATTATCGCTGGATCTTCCCGTCGTAGTGAGCCAGGTGAAATATCTTATGTTCACTTTGCCGTTAACCATCCCCAATTCTCTGAGGAGAATTACGACAAGGATGTGAGCATCGTACGAGTAACACATGCCATCCACTTCGGCCCAAATATCCAGCAGGGTGCTATTATCCAACAAGGTGTCGTAATACCCCAGGGTATTTTTGTTGATCTGCTCGGATGGGGAACTACCGTTCAAGGTGGCAGTGCATCTGACGGCAACCTCCACAAGCTCGAACTCATTGTGACCAACAAAGAAAACTGCAGGAACCAGTATAAGGATCATGATCGCGTCGTGACTGATCACAAATTCTGCGCCGGTTTGGTCCGGGATGGTGGCCGTGACTACGATAATACTGACTTGGGTGCCCCTGCCTTCTTCCAAAATGCGTTGCAAGACTCGGTGCCAGGTCATAAAGCTCGGTCTACGCAGTCTTGGTTGGAAACGAACGTTTCGGACTTCATCAGAGCTGAAGACTGGCCGTCGTCTAGTCCCGATCTTAATCCGCTGGATTATGATTTATGGTCAGTTTTAGAGAGTACGGCTTGCTCTAAACGCCATGATAATTTGGAGTCCCTAAAACAATCCGTACGATTGGCAGTGAACATTTTTCCCGTGGAAAGAGTGCGTGCTTCAATTGATAACTGGCCTCAACGTTTAAAGGACTGTATTGCAGCCAATGGAGACCACTTCGAATAA
Protein
MVSVLTLAVAVLAAVCSCTASNLGRPVSIGEHPSLVQIEVFLPILNQWFQQCAGIVLTNYHYLSTATCFHGEFYDPAYRRIIAGSSRRSEPGEISYVHFAVNHPQFSEENYDKDVSIVRVTHAIHFGPNIQQGAIIQQGVVIPQGIFVDLLGWGTTVQGGSASDGNLHKLELIVTNKENCRNQYKDHDRVVTDHKFCAGLVRDGGRDYDNTDLGAPAFFQNALQDSVPGHKARSTQSWLETNVSDFIRAEDWPSSSPDLNPLDYDLWSVLESTACSKRHDNLESLKQSVRLAVNIFPVERVRASIDNWPQRLKDCIAANGDHFE
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01024727
BABH01024730
DQ443313
KM027249
ABF51402.1
AIU94618.1
+ More
BABH01024734 JN120256 AER70119.1 KZ150010 PZC75085.1 AM690450 CAM84320.1 NWSH01000631 PCG75149.1 ODYU01002467 SOQ39998.1 ODYU01006115 SOQ47675.1 FJ205440 ACR16004.1 KR024671 ALE15213.1 PZC75088.1 GDQN01006776 JAT84278.1 SOQ39997.1 JN793548 AFO68328.1 PCG75147.1 JTDY01003642 KOB69233.1 AY953065 AAX62038.1 PCG75146.1 JQ904129 AFM77759.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JTDY01004416 KOB68185.1 EF600059 ABU98624.1 AY953066 AAX62039.1
BABH01024734 JN120256 AER70119.1 KZ150010 PZC75085.1 AM690450 CAM84320.1 NWSH01000631 PCG75149.1 ODYU01002467 SOQ39998.1 ODYU01006115 SOQ47675.1 FJ205440 ACR16004.1 KR024671 ALE15213.1 PZC75088.1 GDQN01006776 JAT84278.1 SOQ39997.1 JN793548 AFO68328.1 PCG75147.1 JTDY01003642 KOB69233.1 AY953065 AAX62038.1 PCG75146.1 JQ904129 AFM77759.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JTDY01004416 KOB68185.1 EF600059 ABU98624.1 AY953066 AAX62039.1
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
2VU8
E-value=2.94027e-12,
Score=173
Ontologies
GO
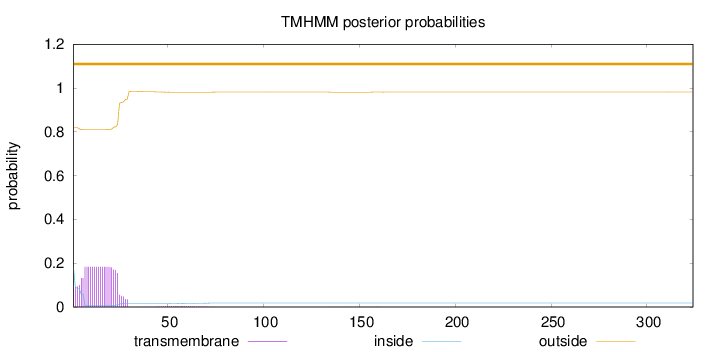
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.980903

Length:
324
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.11437
Exp number, first 60 AAs:
4.07527
Total prob of N-in:
0.18024
outside
1 - 324
Population Genetic Test Statistics
Pi
229.590505
Theta
43.301357
Tajima's D
1.086078
CLR
0.343219
CSRT
0.685815709214539
Interpretation
Uncertain
