Gene
KWMTBOMO03768
Pre Gene Modal
BGIBMGA010061
Annotation
PREDICTED:_trypsin?_alkaline_C-like_[Papilio_xuthus]
Full name
Trypsin, alkaline C
+ More
Trypsin, alkaline A
Trypsin, alkaline B
Trypsin CFT-1
Trypsin, alkaline A
Trypsin, alkaline B
Trypsin CFT-1
Location in the cell
Extracellular Reliability : 3.495
Sequence
CDS
ATGCCGAATTACGAGGTACCGGATAACCAGCCCGTCGTCCACGCCGGCTGGGGGGCCACAAGTCAAGGAGGCGAGGCGTCTGATGTACTTTTGGACGTCACAATCTACACTGTGAACAGGCAGTTATGCGCTGATAGATACTCCACCCTGGGTGATATTATAACCGAGAACATGATTTGCGCTGGTATTTTGGACGTGGGCGGCAAGGACGCCTGCCAAGGTGACTCTGGGGGTCCACTGTATTTCCATAACGTGCTTGTTGGTGTGGTTTCCTGGGGAGAAGGCTGCGCGAACAATACCTTCCCCGGAGTCAGTGCCGCGGTGTCTCCATACACTCAATGGATCGTCGACACGGCTGTTTAA
Protein
MPNYEVPDNQPVVHAGWGATSQGGEASDVLLDVTIYTVNRQLCADRYSTLGDIITENMICAGILDVGGKDACQGDSGGPLYFHNVLVGVVSWGEGCANNTFPGVSAAVSPYTQWIVDTAV
Summary
Catalytic Activity
Preferential cleavage: Arg-|-Xaa, Lys-|-Xaa.
Similarity
Belongs to the peptidase S1 family.
Keywords
Disulfide bond
Hydrolase
Protease
Secreted
Serine protease
Signal
Zymogen
Direct protein sequencing
Feature
propeptide Activation peptide
chain Trypsin, alkaline C
chain Trypsin, alkaline C
Uniprot
H9JKL0
A0A194QJ28
A0A223AIS8
A0A0M4MT18
Q9NB80
Q9NB81
+ More
A0A2W1BP62 A0A2A4JSY0 B1NLE4 Q6R560 A0A2A4JTP3 A0A2W1BJP2 I7C7Q2 Q6T3V9 C9W8J1 A0A1E1WB94 A0A1E1W498 Q4L1L0 A0A2H1VGP3 A0A089RSX5 A0A2H1VGQ2 I7DEJ5 A0A2A4JUT0 A0A194QD40 A5CG75 O45043 A0A2H1W3L1 A0A2H1WEE4 I6TEW4 Q4L1L3 A0A194QET8 Q4L1L1 A0A2W1BTM0 Q9U4I4 B6D1Q8 A0A0L7KYH0 Q9U4I5 A0A212EW56 A0A437BSH3 C9W8I6 A0A0L7L163 Q9NB89 B6CME9 Q4L1L4 A0A223AIT9 A0A0M4M544 A0A2A4JTX8 B4X9A2 A0A345H7U4 Q56IB2 A0A437BTC7 Q56IB0 G5CV08 Q9NB87 Q4L1L5 B1NLD8 Q56IB1 A0A437BG86 Q56IB8 E2IGY7 E0W9N5 Q56IB6 Q6R561 A0A194QDG3 Q56IB4 Q56IB7 Q9TXE6 A0A0N1PGP3 A0A0N0PA32 H9J229 Q56IB5 A0A437BL04 A0A194QD45 H9JV30 J7JYV8 A0A0N1IGP3 S5PVP1 P35047 A0A0L7KUC9 I6SJ30 A0A194PQ27 A0A194PNR3 A0A1E1WDA8 P35045 I6SW38 H9J230 Q961Y0 A0A2W1BE69 O62598 I6TMW8 P35046 A0A2H1VG89 A0A194PQQ5 A0A437BKZ6 H9J243 Q27540 P35042 V9LL27 A0A2H1VJ41 A0A2W1BKF9
A0A2W1BP62 A0A2A4JSY0 B1NLE4 Q6R560 A0A2A4JTP3 A0A2W1BJP2 I7C7Q2 Q6T3V9 C9W8J1 A0A1E1WB94 A0A1E1W498 Q4L1L0 A0A2H1VGP3 A0A089RSX5 A0A2H1VGQ2 I7DEJ5 A0A2A4JUT0 A0A194QD40 A5CG75 O45043 A0A2H1W3L1 A0A2H1WEE4 I6TEW4 Q4L1L3 A0A194QET8 Q4L1L1 A0A2W1BTM0 Q9U4I4 B6D1Q8 A0A0L7KYH0 Q9U4I5 A0A212EW56 A0A437BSH3 C9W8I6 A0A0L7L163 Q9NB89 B6CME9 Q4L1L4 A0A223AIT9 A0A0M4M544 A0A2A4JTX8 B4X9A2 A0A345H7U4 Q56IB2 A0A437BTC7 Q56IB0 G5CV08 Q9NB87 Q4L1L5 B1NLD8 Q56IB1 A0A437BG86 Q56IB8 E2IGY7 E0W9N5 Q56IB6 Q6R561 A0A194QDG3 Q56IB4 Q56IB7 Q9TXE6 A0A0N1PGP3 A0A0N0PA32 H9J229 Q56IB5 A0A437BL04 A0A194QD45 H9JV30 J7JYV8 A0A0N1IGP3 S5PVP1 P35047 A0A0L7KUC9 I6SJ30 A0A194PQ27 A0A194PNR3 A0A1E1WDA8 P35045 I6SW38 H9J230 Q961Y0 A0A2W1BE69 O62598 I6TMW8 P35046 A0A2H1VG89 A0A194PQQ5 A0A437BKZ6 H9J243 Q27540 P35042 V9LL27 A0A2H1VJ41 A0A2W1BKF9
EC Number
3.4.21.4
Pubmed
EMBL
BABH01024725
KQ459167
KPJ03461.1
KX951432
KY684226
ARR75600.1
+ More
ASS33865.1 KR024671 ALE15213.1 AF261982 AAF74744.1 AF261981 AAF74743.1 KZ150010 PZC75087.1 NWSH01000631 PCG75147.1 EF600059 ABU98624.1 AY513650 AAR98919.1 PCG75146.1 PZC75088.1 JN793548 AFO68328.1 AY437836 AAR20817.1 FJ205440 ACR16004.1 GDQN01006776 JAT84278.1 GDQN01009270 JAT81784.1 AY587155 AAT95347.1 ODYU01002467 SOQ39998.1 KM083776 AIR09757.1 SOQ39997.1 JN793549 AFO68329.1 PCG75152.1 KPJ03458.1 AM690450 CAM84320.1 AF045138 AAC02216.1 ODYU01006115 SOQ47675.1 ODYU01007983 SOQ51222.1 JQ904129 AFM77759.1 AY587152 AAT95344.1 KPJ03460.1 AY587154 AAT95346.1 PZC75083.1 AF173498 AAF24228.1 EU673451 ACI45414.1 JTDY01004416 KOB68185.1 AF173497 AAF24227.1 AGBW02012089 OWR45697.1 RSAL01000013 RVE53438.1 FJ205433 ACR15999.2 JTDY01003642 KOB69233.1 AF261973 AAF74735.1 EU325549 ACB54939.1 AY587151 AAT95343.1 KY684221 ASS33860.1 KR024672 ALE15214.1 PCG75148.1 EF531638 ABR88249.1 MF770251 AXG72654.1 AY953065 AAX62038.1 RVE53439.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JN410827 AEP25403.1 AF261975 AAF74737.1 AY587150 AAT95342.1 EF600053 ABU98618.1 PZC75086.1 AY953066 AAX62039.1 RSAL01000066 RVE49371.1 AY953059 AAX62032.1 HM535626 ADK66277.1 FJ205419 ADM35106.1 AY953061 AAX62034.1 AY513649 AAR98918.1 KPJ03459.1 AY953063 AAX62036.1 AY953060 AAX62033.1 KQ460409 KPJ14943.1 KQ458883 KPJ04582.1 BABH01007520 AY953062 AAX62035.1 RSAL01000038 RVE51124.1 KPJ03463.1 BABH01026286 JX312360 AFQ59994.1 KQ459933 KPJ19289.1 KC175562 AGR92345.1 L16807 JTDY01005557 KOB66887.1 JQ904122 AFM77752.1 KQ459597 KPI95068.1 KPI95066.1 GDQN01010607 GDQN01006137 JAT80447.1 JAT84917.1 L16805 JQ904125 AFM77755.1 BABH01007517 AY040819 AAK81696.1 KZ150104 PZC73452.1 AF064525 AF064526 AAC36247.1 AAC36248.1 JQ904123 AFM77753.1 L16806 ODYU01002419 SOQ39873.1 KPI95074.1 RVE51122.1 BABH01007473 U12917 AAA84423.1 L04749 JQ340915 AFP74111.1 ODYU01002655 SOQ40442.1 KZ149992 PZC75529.1
ASS33865.1 KR024671 ALE15213.1 AF261982 AAF74744.1 AF261981 AAF74743.1 KZ150010 PZC75087.1 NWSH01000631 PCG75147.1 EF600059 ABU98624.1 AY513650 AAR98919.1 PCG75146.1 PZC75088.1 JN793548 AFO68328.1 AY437836 AAR20817.1 FJ205440 ACR16004.1 GDQN01006776 JAT84278.1 GDQN01009270 JAT81784.1 AY587155 AAT95347.1 ODYU01002467 SOQ39998.1 KM083776 AIR09757.1 SOQ39997.1 JN793549 AFO68329.1 PCG75152.1 KPJ03458.1 AM690450 CAM84320.1 AF045138 AAC02216.1 ODYU01006115 SOQ47675.1 ODYU01007983 SOQ51222.1 JQ904129 AFM77759.1 AY587152 AAT95344.1 KPJ03460.1 AY587154 AAT95346.1 PZC75083.1 AF173498 AAF24228.1 EU673451 ACI45414.1 JTDY01004416 KOB68185.1 AF173497 AAF24227.1 AGBW02012089 OWR45697.1 RSAL01000013 RVE53438.1 FJ205433 ACR15999.2 JTDY01003642 KOB69233.1 AF261973 AAF74735.1 EU325549 ACB54939.1 AY587151 AAT95343.1 KY684221 ASS33860.1 KR024672 ALE15214.1 PCG75148.1 EF531638 ABR88249.1 MF770251 AXG72654.1 AY953065 AAX62038.1 RVE53439.1 AY953064 AY953068 AAX62037.1 AAX63384.1 JN410827 AEP25403.1 AF261975 AAF74737.1 AY587150 AAT95342.1 EF600053 ABU98618.1 PZC75086.1 AY953066 AAX62039.1 RSAL01000066 RVE49371.1 AY953059 AAX62032.1 HM535626 ADK66277.1 FJ205419 ADM35106.1 AY953061 AAX62034.1 AY513649 AAR98918.1 KPJ03459.1 AY953063 AAX62036.1 AY953060 AAX62033.1 KQ460409 KPJ14943.1 KQ458883 KPJ04582.1 BABH01007520 AY953062 AAX62035.1 RSAL01000038 RVE51124.1 KPJ03463.1 BABH01026286 JX312360 AFQ59994.1 KQ459933 KPJ19289.1 KC175562 AGR92345.1 L16807 JTDY01005557 KOB66887.1 JQ904122 AFM77752.1 KQ459597 KPI95068.1 KPI95066.1 GDQN01010607 GDQN01006137 JAT80447.1 JAT84917.1 L16805 JQ904125 AFM77755.1 BABH01007517 AY040819 AAK81696.1 KZ150104 PZC73452.1 AF064525 AF064526 AAC36247.1 AAC36248.1 JQ904123 AFM77753.1 L16806 ODYU01002419 SOQ39873.1 KPI95074.1 RVE51122.1 BABH01007473 U12917 AAA84423.1 L04749 JQ340915 AFP74111.1 ODYU01002655 SOQ40442.1 KZ149992 PZC75529.1
Proteomes
Pfam
Interpro
IPR033116
TRYPSIN_SER
+ More
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR012224 Pept_S1A_FX
IPR013662 RIH_assoc-dom
IPR000699 RIH_dom
IPR014821 Ins145_P3_rcpt
IPR036300 MIR_dom_sf
IPR035910 RyR/IP3R_RIH_dom_sf
IPR005821 Ion_trans_dom
IPR000493 InsP3_rcpt
IPR016093 MIR_motif
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR018114 TRYPSIN_HIS
IPR012224 Pept_S1A_FX
IPR013662 RIH_assoc-dom
IPR000699 RIH_dom
IPR014821 Ins145_P3_rcpt
IPR036300 MIR_dom_sf
IPR035910 RyR/IP3R_RIH_dom_sf
IPR005821 Ion_trans_dom
IPR000493 InsP3_rcpt
IPR016093 MIR_motif
CDD
ProteinModelPortal
H9JKL0
A0A194QJ28
A0A223AIS8
A0A0M4MT18
Q9NB80
Q9NB81
+ More
A0A2W1BP62 A0A2A4JSY0 B1NLE4 Q6R560 A0A2A4JTP3 A0A2W1BJP2 I7C7Q2 Q6T3V9 C9W8J1 A0A1E1WB94 A0A1E1W498 Q4L1L0 A0A2H1VGP3 A0A089RSX5 A0A2H1VGQ2 I7DEJ5 A0A2A4JUT0 A0A194QD40 A5CG75 O45043 A0A2H1W3L1 A0A2H1WEE4 I6TEW4 Q4L1L3 A0A194QET8 Q4L1L1 A0A2W1BTM0 Q9U4I4 B6D1Q8 A0A0L7KYH0 Q9U4I5 A0A212EW56 A0A437BSH3 C9W8I6 A0A0L7L163 Q9NB89 B6CME9 Q4L1L4 A0A223AIT9 A0A0M4M544 A0A2A4JTX8 B4X9A2 A0A345H7U4 Q56IB2 A0A437BTC7 Q56IB0 G5CV08 Q9NB87 Q4L1L5 B1NLD8 Q56IB1 A0A437BG86 Q56IB8 E2IGY7 E0W9N5 Q56IB6 Q6R561 A0A194QDG3 Q56IB4 Q56IB7 Q9TXE6 A0A0N1PGP3 A0A0N0PA32 H9J229 Q56IB5 A0A437BL04 A0A194QD45 H9JV30 J7JYV8 A0A0N1IGP3 S5PVP1 P35047 A0A0L7KUC9 I6SJ30 A0A194PQ27 A0A194PNR3 A0A1E1WDA8 P35045 I6SW38 H9J230 Q961Y0 A0A2W1BE69 O62598 I6TMW8 P35046 A0A2H1VG89 A0A194PQQ5 A0A437BKZ6 H9J243 Q27540 P35042 V9LL27 A0A2H1VJ41 A0A2W1BKF9
A0A2W1BP62 A0A2A4JSY0 B1NLE4 Q6R560 A0A2A4JTP3 A0A2W1BJP2 I7C7Q2 Q6T3V9 C9W8J1 A0A1E1WB94 A0A1E1W498 Q4L1L0 A0A2H1VGP3 A0A089RSX5 A0A2H1VGQ2 I7DEJ5 A0A2A4JUT0 A0A194QD40 A5CG75 O45043 A0A2H1W3L1 A0A2H1WEE4 I6TEW4 Q4L1L3 A0A194QET8 Q4L1L1 A0A2W1BTM0 Q9U4I4 B6D1Q8 A0A0L7KYH0 Q9U4I5 A0A212EW56 A0A437BSH3 C9W8I6 A0A0L7L163 Q9NB89 B6CME9 Q4L1L4 A0A223AIT9 A0A0M4M544 A0A2A4JTX8 B4X9A2 A0A345H7U4 Q56IB2 A0A437BTC7 Q56IB0 G5CV08 Q9NB87 Q4L1L5 B1NLD8 Q56IB1 A0A437BG86 Q56IB8 E2IGY7 E0W9N5 Q56IB6 Q6R561 A0A194QDG3 Q56IB4 Q56IB7 Q9TXE6 A0A0N1PGP3 A0A0N0PA32 H9J229 Q56IB5 A0A437BL04 A0A194QD45 H9JV30 J7JYV8 A0A0N1IGP3 S5PVP1 P35047 A0A0L7KUC9 I6SJ30 A0A194PQ27 A0A194PNR3 A0A1E1WDA8 P35045 I6SW38 H9J230 Q961Y0 A0A2W1BE69 O62598 I6TMW8 P35046 A0A2H1VG89 A0A194PQQ5 A0A437BKZ6 H9J243 Q27540 P35042 V9LL27 A0A2H1VJ41 A0A2W1BKF9
PDB
6O1G
E-value=1.55409e-19,
Score=229
Ontologies
GO
Topology
Subcellular location
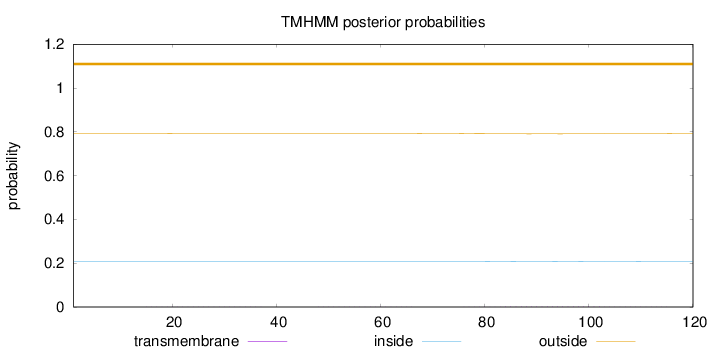
Length:
120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05779
Exp number, first 60 AAs:
0.01068
Total prob of N-in:
0.20762
outside
1 - 120
Population Genetic Test Statistics
Pi
168.13797
Theta
25.959047
Tajima's D
1.015622
CLR
6.824837
CSRT
0.669166541672916
Interpretation
Uncertain
