Gene
KWMTBOMO03766
Pre Gene Modal
BGIBMGA010059
Annotation
transient_receptor_potential_channel_pyrexia-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.057
Sequence
CDS
ATGGACAACTTCGGGTATCGCGAGATGGGGTGGCCTTCACCCTTGCAAACTCCAACAAGATCACGATTAAGCAGCAGCATTCGATCCAGAGGGCGGGAGCCTGAAGAATTTCCACTCAATCCTCAATATAAGCACGTGAAAAACATCGATTTGCTGGAGTCAGCTCGTGAGCCGGGAAAAGCTAAGGAGTACCTTTTCGTCGGACCTTCACCTCCAGCTGACGATGCACCGAACATGTACCACAGTTTCGACGTCCTCGCTCCCGACCCCGATGCTAGATTGACCGATGACGCGGCAAGAAGAGGATTGTACGAAAGAGCCCTCACTCTGGGGGCGGGAAGATTTTTCGATGACTTAGAATGCGGCTTGATAACTGCCGAAAACATCGAGGAACACATTAGTTCTGCTCCTGAAGCGGTCGTAAATTTAACTTTACTATGGGCCTCGTATTTGGCGCGGGATGAGTTATTACCCGGAGTTTTGGACGCTGGGGCGGATATTCACTACTCGGATTCGTCAGGTTTGACTGCCTTACATTTGTCAGCCTTTAGCGGTGCCGGCAGAGCGGCTGTGTTCCTGATATCACGGGGTGCTGACGTCGATTTCGTTCCTAAATATTTCACCCCCCTTCATTGTGCCGCATTCGGGAATTCGTTAGAAGTTGCAGAAATACTGATAGCAAATGGCGCTTCGTTACACGGCGTGGTACAGCGAGCAGGATGCGAAGATAACCTAGTACATTGCGCGGTGAGAGCAGATGCCGTTGAATGTATGGAAATGTTCATAGAACGAGGAGTGGACCCCGCATATGCTACTTCTGGAGGATTGAACGCGTTGCATCTCGCTGCTGAGCTGGGTTCGCGACGTTGCTTGTCGTATTTATTAAGAGAAACAAAGATAAGCGTGAATGGCATGAGTAAACAAAGAGATAAAGAATGTACTGCGTTGCACTTGGCAGCTTCCAGGGGCTATGTGGAGTGCGTAGAACTTTTACTATCTGAAGGTGCGAAAGCAAACACGAAAAATTATAGAGGATTCACGGCCTTACATCTAGCTGCTAGATTATCAAGTATCGACTGCGTAGAAGTGTTGTTGCGAGACGGCAACGCGGACCCGAATGCTGAAGATTACGACAAAAGAAGTCCTTTACACGCTGCGATAACTCGTTCCGAACGTGCTTGTGATATAATCGAACTGCTGGTTAATTGGGGAGCTCAGGTCAACAAGAAAGATGAATATGGGTATTCTGCAATTCATTTAGCTGCAATGGATGGTTTGACGCAATGCGTCGAAACATTAATCTTTCTGGGCGCCGACGTAACTTCGAAATCTAAAAAAGGTCACACAGCGTTAAGTGTTATCGCCAGGAAAACTCCTAAGTCTCTTGCTATTTTAAAACACAACCTAGATTGTGGAATATCATTGAGTCGATCAATAGAAGGCAACGAAGAGGTACAGATCGAATTTGATTTTGCGGTAGTCAAAGCGTGTCAAGGTAAGCATTCGAAGAAATATGGTGTCCCAAATGAATTATGTCAGCCGCAATCAATTTTGGGAGTCATTTTAAACGATAATCCTATCGAATTTGAGAGATGGGTCTTGATGGCAATCACAGTCTTCGAAATCGTTCGAAAATTAACCGGTATAACCGGATACTCGAGTTTCTACCAATATTTTACGACCTTCGAAAACTTGATGGAGTGGTTCGTGCTGCTCTCAGTATTTTCATTGTACAACATTCGCAATGATTACAGCTGGCAGAATCATGTTGGTGGCTATGCTGTTCTTGGGGCTTGGACAAACTTGATGTTGATGATGGGCCAGCTTCCAATGTTTGGCGATTACGTGGCCATGTATCAAAAAGTTTTAATGGAGTTTCTGAAACTTTTATTGGCTTACATTTGTCTACTTCTCGGCTTTACCATTTGCTTCTGTGTCGTCTTTCCTAATGAAGAAATGTTTTCAAATCCTTTGATGGGTTTCATCAGTACTCTTTCGATGATGGTTGGAGAATTAAATTTAAATATCCTAATAAACGATCCGATGCAAGATGATCCTCCTATATTTTTCGAATTGTCAGCACAAATAATTTTCATTTTCTTCCTTATGTTCGTGACTATAATCCTAATGAATCTATTAGTCGGTATTGCGGTTCACGACATCCAAGGATTAAGGAAGACAGCAGGATTATCAAAATTGGTGCGACAGACGAAACTAATTCTTTTTGTTGAAATGGGCATGTTTAGTGCTTGGCTGCCGAAATGCCTACACAAATATGTCTACAGAACAGCTTTAGTATCTCCTGAGGCCGGCAAAGTCATATTAAGCGTGAAACCGTTGAATCCAAGAGAAAAACGTTTACCAACAGACATTATGATGGCAGCTTATGAATTAGCCCAGCTGAACAAAGTTAAGTCCGGCCGATCAGTTAAAGAAGTGCTATACAAGAACAAGATCTCCTCAAAGTTGAAGAACGAAGGACATAATAATGAGCAGAACGTCGGCTTTGAGATTCGAGGAATGCAAGAGAAGATTGACCAAGCGACGTTTAACTTGAAGAAAATAGACCAGGAGATGCGACATTTGAATACTCTTTTAATGGAACAGCAAAATTTCTTTCAAAGTCTCTTCAAGAGCACAGAATTGGTTCCGTACAAATCCCAGCATGCTTCTACCCCAGTTTATTCAGATTCCCCTATCATCTTTGGCAACAATACGTAG
Protein
MDNFGYREMGWPSPLQTPTRSRLSSSIRSRGREPEEFPLNPQYKHVKNIDLLESAREPGKAKEYLFVGPSPPADDAPNMYHSFDVLAPDPDARLTDDAARRGLYERALTLGAGRFFDDLECGLITAENIEEHISSAPEAVVNLTLLWASYLARDELLPGVLDAGADIHYSDSSGLTALHLSAFSGAGRAAVFLISRGADVDFVPKYFTPLHCAAFGNSLEVAEILIANGASLHGVVQRAGCEDNLVHCAVRADAVECMEMFIERGVDPAYATSGGLNALHLAAELGSRRCLSYLLRETKISVNGMSKQRDKECTALHLAASRGYVECVELLLSEGAKANTKNYRGFTALHLAARLSSIDCVEVLLRDGNADPNAEDYDKRSPLHAAITRSERACDIIELLVNWGAQVNKKDEYGYSAIHLAAMDGLTQCVETLIFLGADVTSKSKKGHTALSVIARKTPKSLAILKHNLDCGISLSRSIEGNEEVQIEFDFAVVKACQGKHSKKYGVPNELCQPQSILGVILNDNPIEFERWVLMAITVFEIVRKLTGITGYSSFYQYFTTFENLMEWFVLLSVFSLYNIRNDYSWQNHVGGYAVLGAWTNLMLMMGQLPMFGDYVAMYQKVLMEFLKLLLAYICLLLGFTICFCVVFPNEEMFSNPLMGFISTLSMMVGELNLNILINDPMQDDPPIFFELSAQIIFIFFLMFVTIILMNLLVGIAVHDIQGLRKTAGLSKLVRQTKLILFVEMGMFSAWLPKCLHKYVYRTALVSPEAGKVILSVKPLNPREKRLPTDIMMAAYELAQLNKVKSGRSVKEVLYKNKISSKLKNEGHNNEQNVGFEIRGMQEKIDQATFNLKKIDQEMRHLNTLLMEQQNFFQSLFKSTELVPYKSQHASTPVYSDSPIIFGNNT
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
H9JKK8
W8VT49
A0A0L7KV80
A0A212EW34
A0A0N1PGH9
A0A194QDC0
+ More
A0A2W1BQS4 A0A2A4JSX4 A0A212EY44 A0A0N1INX4 A0A194QDF8 A0A2A4J425 A0A2H1VVM8 W8VYR7 H9JKH6 A0A2W1BN88 A0A0L7KRS9 A0A182FIN2 W5JSU2 A0A084VFU2 A0A182IYC8 A0A1W4XF69 A0A1J1IHM1 A0A182F493 B5X0K8 A0A1J1IG66 A0A1W4VYY7 Q6NQV8 B3NYS8 Q9VHY7 B4R0A9 B4I4Y3 A0A2M4CRT3 A0A2M4ACA0 A0A084W5L3 A0A1W4VZM1 A0A2M4CRM4 A0A2M4ACK1 A0A0B4LGT2 B4PRD7 A0A0R1E1C4 A0A182QEN6 A0A182JF65 A0A0R1E1Q9 A0A182N8V8 A0A1Y1KNZ5 A0A1S4H4F7 Q7Q0M1 A0A182XRE9 A0A182IGN3 A0A182S7D4 A0A182VST2 A0A182MKC6 A0A182RAE2 A0A182YN03 A0A182JSA4 A0A182MF93 A0A139WKY6 A0A182R283 A0A182W659 A0A182VHX2 Q7QF89 A0A182KA14 A0A182WTJ0 A0A182UKQ4 A0A182IAZ3 A0A182N0Q4 A0A182S924 A0A182QYF5 A0A182PBH5 A0A182YF07 A0A1L8DK70 A0A0T6B2H6 N6TX17 U4U6Z5 A0A2M4AF87 A0A067RHV7 B0WRI4 A0A1B6LIV3 A0A1Q3F007 A0A1B6F4A2 A0A1B6JM30 A0A182PPV1 T1H7Z3 A0A023EXA0 A0A067QUF4 A0A0K8TDM4 A0A0C9QZ02 A0A0C9QQC9
A0A2W1BQS4 A0A2A4JSX4 A0A212EY44 A0A0N1INX4 A0A194QDF8 A0A2A4J425 A0A2H1VVM8 W8VYR7 H9JKH6 A0A2W1BN88 A0A0L7KRS9 A0A182FIN2 W5JSU2 A0A084VFU2 A0A182IYC8 A0A1W4XF69 A0A1J1IHM1 A0A182F493 B5X0K8 A0A1J1IG66 A0A1W4VYY7 Q6NQV8 B3NYS8 Q9VHY7 B4R0A9 B4I4Y3 A0A2M4CRT3 A0A2M4ACA0 A0A084W5L3 A0A1W4VZM1 A0A2M4CRM4 A0A2M4ACK1 A0A0B4LGT2 B4PRD7 A0A0R1E1C4 A0A182QEN6 A0A182JF65 A0A0R1E1Q9 A0A182N8V8 A0A1Y1KNZ5 A0A1S4H4F7 Q7Q0M1 A0A182XRE9 A0A182IGN3 A0A182S7D4 A0A182VST2 A0A182MKC6 A0A182RAE2 A0A182YN03 A0A182JSA4 A0A182MF93 A0A139WKY6 A0A182R283 A0A182W659 A0A182VHX2 Q7QF89 A0A182KA14 A0A182WTJ0 A0A182UKQ4 A0A182IAZ3 A0A182N0Q4 A0A182S924 A0A182QYF5 A0A182PBH5 A0A182YF07 A0A1L8DK70 A0A0T6B2H6 N6TX17 U4U6Z5 A0A2M4AF87 A0A067RHV7 B0WRI4 A0A1B6LIV3 A0A1Q3F007 A0A1B6F4A2 A0A1B6JM30 A0A182PPV1 T1H7Z3 A0A023EXA0 A0A067QUF4 A0A0K8TDM4 A0A0C9QZ02 A0A0C9QQC9
Pubmed
EMBL
BABH01024724
AB703647
BAO53212.1
JTDY01005497
KOB66934.1
AGBW02012089
+ More
OWR45696.1 KQ460409 KPJ14946.1 KQ459167 KPJ03457.1 KZ150010 PZC75090.1 NWSH01000631 PCG75145.1 AGBW02011619 OWR46419.1 KQ460938 KPJ10658.1 KPJ03454.1 NWSH01003455 PCG66264.1 ODYU01004410 SOQ44254.1 AB437369 BAO53210.1 BABH01024703 PZC75094.1 JTDY01006675 KOB65795.1 ADMH02000465 ETN66368.1 ATLV01012463 KE524793 KFB36836.1 CVRI01000047 CRK98534.1 BT044577 ACI16539.1 CRK98532.1 BT010330 AAQ55961.1 CH954181 EDV48191.1 AE014297 BT003789 AAF54159.2 AAF54160.2 AAO41472.1 AAS65124.1 CM000364 EDX12034.1 CH480821 EDW55276.1 GGFL01003807 MBW67985.1 GGFK01005095 MBW38416.1 ATLV01020673 KE525304 KFB45507.1 GGFL01003806 MBW67984.1 GGFK01005204 MBW38525.1 AHN57213.1 CM000160 EDW96325.1 KRK03078.1 AXCN02001867 KRK03077.1 GEZM01081070 JAV61910.1 AAAB01008980 EAA13954.3 APCN01005275 AXCM01002492 AXCM01003740 KQ971323 KYB28553.1 AAAB01008846 EAA06325.5 APCN01001381 AXCN02001398 GFDF01007248 JAV06836.1 LJIG01016189 KRT81349.1 APGK01051763 APGK01051764 APGK01051765 KB741205 ENN72936.1 KB631811 ERL86356.1 GGFK01006123 MBW39444.1 KK852498 KDR22603.1 DS232056 EDS33382.1 GEBQ01016361 GEBQ01004306 JAT23616.1 JAT35671.1 GFDL01014150 JAV20895.1 GECZ01024751 JAS45018.1 GECU01007418 JAT00289.1 ACPB03012378 GBBI01004900 JAC13812.1 KK852942 KDR13512.1 GBRD01002148 GDHC01003903 JAG63673.1 JAQ14726.1 GBYB01005912 JAG75679.1 GBYB01005909 JAG75676.1
OWR45696.1 KQ460409 KPJ14946.1 KQ459167 KPJ03457.1 KZ150010 PZC75090.1 NWSH01000631 PCG75145.1 AGBW02011619 OWR46419.1 KQ460938 KPJ10658.1 KPJ03454.1 NWSH01003455 PCG66264.1 ODYU01004410 SOQ44254.1 AB437369 BAO53210.1 BABH01024703 PZC75094.1 JTDY01006675 KOB65795.1 ADMH02000465 ETN66368.1 ATLV01012463 KE524793 KFB36836.1 CVRI01000047 CRK98534.1 BT044577 ACI16539.1 CRK98532.1 BT010330 AAQ55961.1 CH954181 EDV48191.1 AE014297 BT003789 AAF54159.2 AAF54160.2 AAO41472.1 AAS65124.1 CM000364 EDX12034.1 CH480821 EDW55276.1 GGFL01003807 MBW67985.1 GGFK01005095 MBW38416.1 ATLV01020673 KE525304 KFB45507.1 GGFL01003806 MBW67984.1 GGFK01005204 MBW38525.1 AHN57213.1 CM000160 EDW96325.1 KRK03078.1 AXCN02001867 KRK03077.1 GEZM01081070 JAV61910.1 AAAB01008980 EAA13954.3 APCN01005275 AXCM01002492 AXCM01003740 KQ971323 KYB28553.1 AAAB01008846 EAA06325.5 APCN01001381 AXCN02001398 GFDF01007248 JAV06836.1 LJIG01016189 KRT81349.1 APGK01051763 APGK01051764 APGK01051765 KB741205 ENN72936.1 KB631811 ERL86356.1 GGFK01006123 MBW39444.1 KK852498 KDR22603.1 DS232056 EDS33382.1 GEBQ01016361 GEBQ01004306 JAT23616.1 JAT35671.1 GFDL01014150 JAV20895.1 GECZ01024751 JAS45018.1 GECU01007418 JAT00289.1 ACPB03012378 GBBI01004900 JAC13812.1 KK852942 KDR13512.1 GBRD01002148 GDHC01003903 JAG63673.1 JAQ14726.1 GBYB01005912 JAG75679.1 GBYB01005909 JAG75676.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053240
UP000053268
UP000218220
+ More
UP000069272 UP000000673 UP000030765 UP000075880 UP000192223 UP000183832 UP000192221 UP000008711 UP000000803 UP000000304 UP000001292 UP000002282 UP000075886 UP000075884 UP000007062 UP000076407 UP000075840 UP000075901 UP000075920 UP000075883 UP000075900 UP000076408 UP000075881 UP000007266 UP000075903 UP000075902 UP000075885 UP000019118 UP000030742 UP000027135 UP000002320 UP000015103
UP000069272 UP000000673 UP000030765 UP000075880 UP000192223 UP000183832 UP000192221 UP000008711 UP000000803 UP000000304 UP000001292 UP000002282 UP000075886 UP000075884 UP000007062 UP000076407 UP000075840 UP000075901 UP000075920 UP000075883 UP000075900 UP000076408 UP000075881 UP000007266 UP000075903 UP000075902 UP000075885 UP000019118 UP000030742 UP000027135 UP000002320 UP000015103
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JKK8
W8VT49
A0A0L7KV80
A0A212EW34
A0A0N1PGH9
A0A194QDC0
+ More
A0A2W1BQS4 A0A2A4JSX4 A0A212EY44 A0A0N1INX4 A0A194QDF8 A0A2A4J425 A0A2H1VVM8 W8VYR7 H9JKH6 A0A2W1BN88 A0A0L7KRS9 A0A182FIN2 W5JSU2 A0A084VFU2 A0A182IYC8 A0A1W4XF69 A0A1J1IHM1 A0A182F493 B5X0K8 A0A1J1IG66 A0A1W4VYY7 Q6NQV8 B3NYS8 Q9VHY7 B4R0A9 B4I4Y3 A0A2M4CRT3 A0A2M4ACA0 A0A084W5L3 A0A1W4VZM1 A0A2M4CRM4 A0A2M4ACK1 A0A0B4LGT2 B4PRD7 A0A0R1E1C4 A0A182QEN6 A0A182JF65 A0A0R1E1Q9 A0A182N8V8 A0A1Y1KNZ5 A0A1S4H4F7 Q7Q0M1 A0A182XRE9 A0A182IGN3 A0A182S7D4 A0A182VST2 A0A182MKC6 A0A182RAE2 A0A182YN03 A0A182JSA4 A0A182MF93 A0A139WKY6 A0A182R283 A0A182W659 A0A182VHX2 Q7QF89 A0A182KA14 A0A182WTJ0 A0A182UKQ4 A0A182IAZ3 A0A182N0Q4 A0A182S924 A0A182QYF5 A0A182PBH5 A0A182YF07 A0A1L8DK70 A0A0T6B2H6 N6TX17 U4U6Z5 A0A2M4AF87 A0A067RHV7 B0WRI4 A0A1B6LIV3 A0A1Q3F007 A0A1B6F4A2 A0A1B6JM30 A0A182PPV1 T1H7Z3 A0A023EXA0 A0A067QUF4 A0A0K8TDM4 A0A0C9QZ02 A0A0C9QQC9
A0A2W1BQS4 A0A2A4JSX4 A0A212EY44 A0A0N1INX4 A0A194QDF8 A0A2A4J425 A0A2H1VVM8 W8VYR7 H9JKH6 A0A2W1BN88 A0A0L7KRS9 A0A182FIN2 W5JSU2 A0A084VFU2 A0A182IYC8 A0A1W4XF69 A0A1J1IHM1 A0A182F493 B5X0K8 A0A1J1IG66 A0A1W4VYY7 Q6NQV8 B3NYS8 Q9VHY7 B4R0A9 B4I4Y3 A0A2M4CRT3 A0A2M4ACA0 A0A084W5L3 A0A1W4VZM1 A0A2M4CRM4 A0A2M4ACK1 A0A0B4LGT2 B4PRD7 A0A0R1E1C4 A0A182QEN6 A0A182JF65 A0A0R1E1Q9 A0A182N8V8 A0A1Y1KNZ5 A0A1S4H4F7 Q7Q0M1 A0A182XRE9 A0A182IGN3 A0A182S7D4 A0A182VST2 A0A182MKC6 A0A182RAE2 A0A182YN03 A0A182JSA4 A0A182MF93 A0A139WKY6 A0A182R283 A0A182W659 A0A182VHX2 Q7QF89 A0A182KA14 A0A182WTJ0 A0A182UKQ4 A0A182IAZ3 A0A182N0Q4 A0A182S924 A0A182QYF5 A0A182PBH5 A0A182YF07 A0A1L8DK70 A0A0T6B2H6 N6TX17 U4U6Z5 A0A2M4AF87 A0A067RHV7 B0WRI4 A0A1B6LIV3 A0A1Q3F007 A0A1B6F4A2 A0A1B6JM30 A0A182PPV1 T1H7Z3 A0A023EXA0 A0A067QUF4 A0A0K8TDM4 A0A0C9QZ02 A0A0C9QQC9
PDB
6MOL
E-value=5.61877e-37,
Score=390
Ontologies
GO
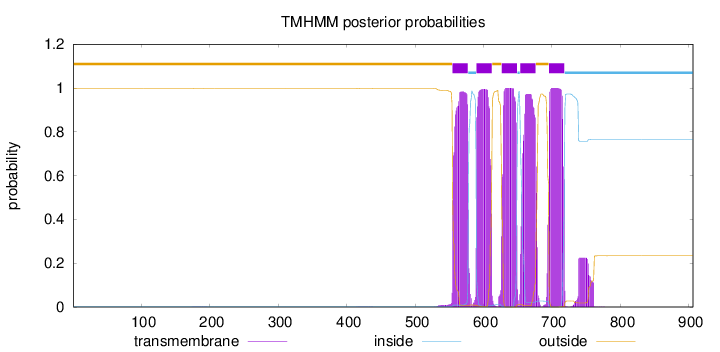
Topology
Length:
906
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
114.16498
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00241
outside
1 - 554
TMhelix
555 - 577
inside
578 - 589
TMhelix
590 - 612
outside
613 - 626
TMhelix
627 - 649
inside
650 - 653
TMhelix
654 - 676
outside
677 - 695
TMhelix
696 - 718
inside
719 - 906
Population Genetic Test Statistics
Pi
181.033771
Theta
177.458795
Tajima's D
0.050751
CLR
1.827332
CSRT
0.380080995950203
Interpretation
Uncertain
