Gene
KWMTBOMO03763
Pre Gene Modal
BGIBMGA010027
Annotation
transient_receptor_potential_channel_pyrexia_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.133
Sequence
CDS
ATGCCGAACACGCGTCGTCTGTTGCCCGCGCGGTGGTTGCGAGCTCGCGCGCCCGATGAGCCGCGGCCCGCGGACCGGCGCGGGCGGGACCTGCCCGACCACGAGTCCCTCGAGCGCGCCTACCCCTCCATGGGGCACCTCGAGTACGTGCTGGCCGGCTCCTCGCCGCCGGCCGAGAGCGCACCCAATATGTACGACAGCTTCGAGGAGCCACCTCTGGACCTGACCGCGCACATCTGTGCCGACTCACTGCGTCAGAGCGCCCACGAGCAAATGCGAGCCGTCGGCGGGCGGTTGCGCCTCCTTGATGAGCTCGAGAGTGGAGTCATTCCAGCTGACGCGACGACTGACGCCTTCGCGACCGCCACCGATGCCGAAAAGAACGTCTGCTTATTTTGGACCGCGTTTTTGAAACTCGCCAACTTGCTACCACCGCTGATTGAGGTTGGGGCGGACCCCCTGTACTACGACTCTCTGGGCCTGTCTCCGCTGCACGTGGCCGCGTTCAGTGGTTCCGTAGAATGCGCCAGTTATCTGCTGTCGTGCGGCGCGGACCCTAATTATATGCCCCGGTGTTTTGCGCCGCTTCACTGTGCCGCATTCGGTAACTCGGTACAAGTGGCTAATTTGCTAATAGCTCGCGGTGCGTCCGTGCACGCGGCTGTAAAGTACATCAATTGCGAAGGAGGTCTCCTTCACTGTGCGGTGCGAGCTAATTCCGCGGAGTGCCTCAAATTGTTTATATCGCACGGAGTTGACGTGAACTTGATCGAGCCGGGCGGTACTAATGCCATTCACCTCGCCGCTGATTTGGGCGTTAAACAGTGTCTTATAATATTGCTTGACACGCCCGGAGCCGACCCTAATGTTAGAACAAGAGTAGGGGACAGAGAATCCACCGCTTTGCATTTAGCCGCCGACGGTGGCTTTGTTGAGTGTGTCGATTTGTTACTCAGCAAAGGTGCAGATGCGAGTTTGAAGAATCATAGGGGGTTCACAGCGTTACATTTGGCAGCTCGGTCTGCGAGTTTAGAATGCGTGGAATCCTTACTCCGGAAAGGCAATGCCGACCCCAACGCTATGGATTTTGATAAACGAACACCGCTACATGCAGCTATCGGAAAATCGGATTCCGCTTGTGATATCATCGAAACCTTAATCAGTTGGGGCGCCAACGTAAACCAAAAAGATGAATACGGATTTACTCCACTGCACCTAGCCGCACTTGACGGACTGTCGGCCTGCGTGGAGACCTTGATATATCACGGTGCTGACGTCACAACGAGATCTAAGAAGGGAAACTCGGCCCTTAATGTAATCGCTAGAAAAACGCCCGCTTCACTCGCGATGGTAACTAGAAAATTGGACTGCGCCATTACTCTACATCACTCGCAGTCGAGCAATAGAGAAGTTGAATTGGAGCTTGATTTTCGTAGCATATTGCAGCATTGTTATCCGCGAGAGATCAGCTATTTGAATACGTTCGTCGACGAAGGCCAAAAAGAAGTCCTACTTCACCCACTGTGCTCGGCCTTTCTGTACATTAAATGGGAGAAAATACCACTCGCGCACAATTGTTATAACGGTAGCAAGGACATGGAGGAGACAATACAAGAGCAGGAGTTGTGTCAAAAGCAATCCATCCTTGGTGACCTATTAAGGAAAAATCCATTCGTAATTGAAATGCAATGGTGGGTTCTAGCCGGGATAACGATATTTGAGATATTTAGGAAGGTATACGGGATCGCTGGATATTCGACCGTCAAACAGTATTTGATGCAATCGGAGAACATTATTGAGTGGTTCGTGATCATTAGTGTTTTCCTAATTTCGTATATATACACAAACATCACGTATACATGGCAGAACCACGTCGGCGCTTTCGCGGTGCTCGCTGGGTGGACGAACCTCATGATGATGATCGGACAGCTACCCGTTTTCGGCACCTACGTCGCGATGTACCAAAAAGTACAAAAGGAATTTGCCAAATTGCTTATGGCCTATTCGTGTATCCTAATCGGTTTCACGATAAGTTTCTGCGTTATCTTCCCGGACTCGTCCTCGTTTGCGAATCCGTTTATGGGCTTCATAACCGTCTTGACAATGATGATCGGGGAGCTAAATTTAGATTTACTACTCAACGAGCCAGATGGAAACGACCCTCCTGTCTTGCTCGAGTTTTCTGCGCAGATAACCTACGTGTTATTCCTAATGTTTGTTACGGTCGTATTGATGAATTTACTGGTCGGCATAGCGGTCCACGATATTCAGGGTTTGAGGAAAACCGCTGGCCTTTCAAAGCTGGTTCGACAAACCAAACTGATATCTTACATGGAATTGGCGTTGTTCAATGGATATTTACCGAAATGTCTACTAAAGATACTACATTCTTCAGCTTTGGTTTCGCCCCAGGCCTACAGGGTTGTGCTAAGCGTCAAACCGTTGAACCCAAGCGAGAAGAGACTGCCTAGGGACATAATGATGGCCGCTTACGACATAGCAAAGATGCGGAAGCAATACGGACACACTATATCATCGAGCGGTTCGACAACGGGCGCCTATTCATGCTTCAAAAAGTACGAGAGCAATAACGACTCCGGGTACCGAGAATACGGTTACTCAGGACTCGGAAGCATCCACGCCAGACTCGACGAGACATCGGAACACGTCAGACAATTGACACAGGAAGTCAAGGAATTGAAGAAGCTAATAAACGCGCAGCAACTTGTCATTCAGCAAGCGCTGGTCGGCGCGATGGACCGTCCGTGA
Protein
MPNTRRLLPARWLRARAPDEPRPADRRGRDLPDHESLERAYPSMGHLEYVLAGSSPPAESAPNMYDSFEEPPLDLTAHICADSLRQSAHEQMRAVGGRLRLLDELESGVIPADATTDAFATATDAEKNVCLFWTAFLKLANLLPPLIEVGADPLYYDSLGLSPLHVAAFSGSVECASYLLSCGADPNYMPRCFAPLHCAAFGNSVQVANLLIARGASVHAAVKYINCEGGLLHCAVRANSAECLKLFISHGVDVNLIEPGGTNAIHLAADLGVKQCLIILLDTPGADPNVRTRVGDRESTALHLAADGGFVECVDLLLSKGADASLKNHRGFTALHLAARSASLECVESLLRKGNADPNAMDFDKRTPLHAAIGKSDSACDIIETLISWGANVNQKDEYGFTPLHLAALDGLSACVETLIYHGADVTTRSKKGNSALNVIARKTPASLAMVTRKLDCAITLHHSQSSNREVELELDFRSILQHCYPREISYLNTFVDEGQKEVLLHPLCSAFLYIKWEKIPLAHNCYNGSKDMEETIQEQELCQKQSILGDLLRKNPFVIEMQWWVLAGITIFEIFRKVYGIAGYSTVKQYLMQSENIIEWFVIISVFLISYIYTNITYTWQNHVGAFAVLAGWTNLMMMIGQLPVFGTYVAMYQKVQKEFAKLLMAYSCILIGFTISFCVIFPDSSSFANPFMGFITVLTMMIGELNLDLLLNEPDGNDPPVLLEFSAQITYVLFLMFVTVVLMNLLVGIAVHDIQGLRKTAGLSKLVRQTKLISYMELALFNGYLPKCLLKILHSSALVSPQAYRVVLSVKPLNPSEKRLPRDIMMAAYDIAKMRKQYGHTISSSGSTTGAYSCFKKYESNNDSGYREYGYSGLGSIHARLDETSEHVRQLTQEVKELKKLINAQQLVIQQALVGAMDRP
Summary
Similarity
Belongs to the transient receptor (TC 1.A.4) family.
Uniprot
H9JKH6
W8VYR7
A0A212EY44
A0A0N1INX4
A0A2A4J425
A0A2H1VVM8
+ More
A0A2W1BN88 A0A194QDF8 A0A0L7KRS9 A0A0L0CKM1 A0A0A1WRK9 A0A0A1WL94 A0A1A9WFM6 A0A1B0FNB9 A0A1A9YI35 A0A1A9Z6J3 B4K7I8 W8BFL5 A0A034W680 A0A0Q9XA97 A0A0K8VD15 A0A034W4P9 A0A1B0GQ57 A0A1B0BY82 A0A1I8MKT5 A0A3B0KG84 A0A3B0K7L8 A0A1I8PRH0 A0A1I8MKS7 A0A0R3NJS7 B4GMX9 A0A1I8PRL8 A0A1I8MKT1 Q294L4 A0A3B0JSR8 A0A3B0JUI7 A0A0R3NJU9 A0A0R3NJP8 I5APS4 A0A1I8PRL4 A0A336LYY4 A0A0M4EH70 B4N8D0 A0A1B0GJI4 B4JI37 B4M0V1 A0A0Q9WUI7 A0A0Q9WSR2 A0A0Q9WIQ4 B3LXF7 A0A0P9C947 A0A0P8YEQ8 B4R0A9 B4I4Y3 B5X0K8 Q9VHY7 Q6NQV8 B3NYS8 B4PRD7 A0A0B4LGT2 A0A0R1E1C4 A0A0R1E1Q9 A0A1W4VYY7 A0A1W4VZM1 A0A1B0CSF7 A0A1B0CSY4 A0A1L8DIW8 A0A1B0GQ56 A0A1L8DIP3 A0A1J1IHM1 A0A1J1IG66 B0WRI4 A0A1Q3F007 A0A182QEN6 A0A182JF65 A0A182N8V8 Q7Q0M1 A0A084W5L3 A0A2M4ACK1 A0A182F493 A0A2M4ACA0 A0A2M4CRT3 A0A2M4CRM4 W5JSU2 A0A1S4H4F7 A0A182IYC8 A0A182FIN2 A0A182N0Q4 A0A084VFU2 A0A182PBH5 A0A182R283 A0A182YF07 A0A182W659 Q7QF89 A0A182MF93 A0A182XRE9 A0A182VHX2 A0A182IGN3 A0A182KA14 A0A182S924
A0A2W1BN88 A0A194QDF8 A0A0L7KRS9 A0A0L0CKM1 A0A0A1WRK9 A0A0A1WL94 A0A1A9WFM6 A0A1B0FNB9 A0A1A9YI35 A0A1A9Z6J3 B4K7I8 W8BFL5 A0A034W680 A0A0Q9XA97 A0A0K8VD15 A0A034W4P9 A0A1B0GQ57 A0A1B0BY82 A0A1I8MKT5 A0A3B0KG84 A0A3B0K7L8 A0A1I8PRH0 A0A1I8MKS7 A0A0R3NJS7 B4GMX9 A0A1I8PRL8 A0A1I8MKT1 Q294L4 A0A3B0JSR8 A0A3B0JUI7 A0A0R3NJU9 A0A0R3NJP8 I5APS4 A0A1I8PRL4 A0A336LYY4 A0A0M4EH70 B4N8D0 A0A1B0GJI4 B4JI37 B4M0V1 A0A0Q9WUI7 A0A0Q9WSR2 A0A0Q9WIQ4 B3LXF7 A0A0P9C947 A0A0P8YEQ8 B4R0A9 B4I4Y3 B5X0K8 Q9VHY7 Q6NQV8 B3NYS8 B4PRD7 A0A0B4LGT2 A0A0R1E1C4 A0A0R1E1Q9 A0A1W4VYY7 A0A1W4VZM1 A0A1B0CSF7 A0A1B0CSY4 A0A1L8DIW8 A0A1B0GQ56 A0A1L8DIP3 A0A1J1IHM1 A0A1J1IG66 B0WRI4 A0A1Q3F007 A0A182QEN6 A0A182JF65 A0A182N8V8 Q7Q0M1 A0A084W5L3 A0A2M4ACK1 A0A182F493 A0A2M4ACA0 A0A2M4CRT3 A0A2M4CRM4 W5JSU2 A0A1S4H4F7 A0A182IYC8 A0A182FIN2 A0A182N0Q4 A0A084VFU2 A0A182PBH5 A0A182R283 A0A182YF07 A0A182W659 Q7QF89 A0A182MF93 A0A182XRE9 A0A182VHX2 A0A182IGN3 A0A182KA14 A0A182S924
Pubmed
19121390
24639527
22118469
26354079
28756777
26227816
+ More
26108605 25830018 17994087 24495485 25348373 25315136 15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 24438588 20920257 23761445 25244985 14747013 17210077
26108605 25830018 17994087 24495485 25348373 25315136 15632085 23185243 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 12364791 24438588 20920257 23761445 25244985 14747013 17210077
EMBL
BABH01024703
AB437369
BAO53210.1
AGBW02011619
OWR46419.1
KQ460938
+ More
KPJ10658.1 NWSH01003455 PCG66264.1 ODYU01004410 SOQ44254.1 KZ150010 PZC75094.1 KQ459167 KPJ03454.1 JTDY01006675 KOB65795.1 JRES01000266 KNC32816.1 GBXI01013142 GBXI01012634 GBXI01011050 JAD01150.1 JAD01658.1 JAD03242.1 GBXI01015114 JAC99177.1 CCAG010014739 CH933806 EDW15332.1 GAMC01014489 GAMC01014488 JAB92066.1 GAKP01009679 JAC49273.1 KRG01518.1 GDHF01015568 GDHF01006261 JAI36746.1 JAI46053.1 GAKP01009680 JAC49272.1 AJVK01006456 JXJN01022512 JXJN01022513 OUUW01000008 SPP84061.1 SPP84060.1 CM000070 KRT01192.1 KRT01193.1 CH479185 EDW38203.1 EAL28950.1 SPP84063.1 SPP84062.1 KRT01191.1 KRT01194.1 EIM52959.2 UFQS01000320 UFQT01000320 SSX02826.1 SSX23194.1 CP012526 ALC47866.1 CH964232 EDW81381.1 AJWK01021796 CH916369 EDV92918.1 CH940650 EDW68410.1 KRF83827.1 KRF83829.1 KRF83828.1 KRF83830.1 CH902617 EDV42801.1 KPU79926.1 KPU79928.1 KPU79925.1 KPU79927.1 CM000364 EDX12034.1 CH480821 EDW55276.1 BT044577 ACI16539.1 AE014297 BT003789 AAF54159.2 AAF54160.2 AAO41472.1 AAS65124.1 BT010330 AAQ55961.1 CH954181 EDV48191.1 CM000160 EDW96325.1 AHN57213.1 KRK03078.1 KRK03077.1 AJWK01026016 AJWK01026773 GFDF01007784 JAV06300.1 AJVK01006453 GFDF01007783 JAV06301.1 CVRI01000047 CRK98534.1 CRK98532.1 DS232056 EDS33382.1 GFDL01014150 JAV20895.1 AXCN02001867 AAAB01008980 EAA13954.3 ATLV01020673 KE525304 KFB45507.1 GGFK01005204 MBW38525.1 GGFK01005095 MBW38416.1 GGFL01003807 MBW67985.1 GGFL01003806 MBW67984.1 ADMH02000465 ETN66368.1 ATLV01012463 KE524793 KFB36836.1 AAAB01008846 EAA06325.5 AXCM01003740 APCN01005275
KPJ10658.1 NWSH01003455 PCG66264.1 ODYU01004410 SOQ44254.1 KZ150010 PZC75094.1 KQ459167 KPJ03454.1 JTDY01006675 KOB65795.1 JRES01000266 KNC32816.1 GBXI01013142 GBXI01012634 GBXI01011050 JAD01150.1 JAD01658.1 JAD03242.1 GBXI01015114 JAC99177.1 CCAG010014739 CH933806 EDW15332.1 GAMC01014489 GAMC01014488 JAB92066.1 GAKP01009679 JAC49273.1 KRG01518.1 GDHF01015568 GDHF01006261 JAI36746.1 JAI46053.1 GAKP01009680 JAC49272.1 AJVK01006456 JXJN01022512 JXJN01022513 OUUW01000008 SPP84061.1 SPP84060.1 CM000070 KRT01192.1 KRT01193.1 CH479185 EDW38203.1 EAL28950.1 SPP84063.1 SPP84062.1 KRT01191.1 KRT01194.1 EIM52959.2 UFQS01000320 UFQT01000320 SSX02826.1 SSX23194.1 CP012526 ALC47866.1 CH964232 EDW81381.1 AJWK01021796 CH916369 EDV92918.1 CH940650 EDW68410.1 KRF83827.1 KRF83829.1 KRF83828.1 KRF83830.1 CH902617 EDV42801.1 KPU79926.1 KPU79928.1 KPU79925.1 KPU79927.1 CM000364 EDX12034.1 CH480821 EDW55276.1 BT044577 ACI16539.1 AE014297 BT003789 AAF54159.2 AAF54160.2 AAO41472.1 AAS65124.1 BT010330 AAQ55961.1 CH954181 EDV48191.1 CM000160 EDW96325.1 AHN57213.1 KRK03078.1 KRK03077.1 AJWK01026016 AJWK01026773 GFDF01007784 JAV06300.1 AJVK01006453 GFDF01007783 JAV06301.1 CVRI01000047 CRK98534.1 CRK98532.1 DS232056 EDS33382.1 GFDL01014150 JAV20895.1 AXCN02001867 AAAB01008980 EAA13954.3 ATLV01020673 KE525304 KFB45507.1 GGFK01005204 MBW38525.1 GGFK01005095 MBW38416.1 GGFL01003807 MBW67985.1 GGFL01003806 MBW67984.1 ADMH02000465 ETN66368.1 ATLV01012463 KE524793 KFB36836.1 AAAB01008846 EAA06325.5 AXCM01003740 APCN01005275
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000053268
UP000037510
+ More
UP000037069 UP000091820 UP000092444 UP000092443 UP000092445 UP000009192 UP000092462 UP000092460 UP000095301 UP000268350 UP000095300 UP000001819 UP000008744 UP000092553 UP000007798 UP000092461 UP000001070 UP000008792 UP000007801 UP000000304 UP000001292 UP000000803 UP000008711 UP000002282 UP000192221 UP000183832 UP000002320 UP000075886 UP000075880 UP000075884 UP000007062 UP000030765 UP000069272 UP000000673 UP000075885 UP000075900 UP000076408 UP000075920 UP000075883 UP000076407 UP000075903 UP000075840 UP000075881 UP000075901
UP000037069 UP000091820 UP000092444 UP000092443 UP000092445 UP000009192 UP000092462 UP000092460 UP000095301 UP000268350 UP000095300 UP000001819 UP000008744 UP000092553 UP000007798 UP000092461 UP000001070 UP000008792 UP000007801 UP000000304 UP000001292 UP000000803 UP000008711 UP000002282 UP000192221 UP000183832 UP000002320 UP000075886 UP000075880 UP000075884 UP000007062 UP000030765 UP000069272 UP000000673 UP000075885 UP000075900 UP000076408 UP000075920 UP000075883 UP000076407 UP000075903 UP000075840 UP000075881 UP000075901
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JKH6
W8VYR7
A0A212EY44
A0A0N1INX4
A0A2A4J425
A0A2H1VVM8
+ More
A0A2W1BN88 A0A194QDF8 A0A0L7KRS9 A0A0L0CKM1 A0A0A1WRK9 A0A0A1WL94 A0A1A9WFM6 A0A1B0FNB9 A0A1A9YI35 A0A1A9Z6J3 B4K7I8 W8BFL5 A0A034W680 A0A0Q9XA97 A0A0K8VD15 A0A034W4P9 A0A1B0GQ57 A0A1B0BY82 A0A1I8MKT5 A0A3B0KG84 A0A3B0K7L8 A0A1I8PRH0 A0A1I8MKS7 A0A0R3NJS7 B4GMX9 A0A1I8PRL8 A0A1I8MKT1 Q294L4 A0A3B0JSR8 A0A3B0JUI7 A0A0R3NJU9 A0A0R3NJP8 I5APS4 A0A1I8PRL4 A0A336LYY4 A0A0M4EH70 B4N8D0 A0A1B0GJI4 B4JI37 B4M0V1 A0A0Q9WUI7 A0A0Q9WSR2 A0A0Q9WIQ4 B3LXF7 A0A0P9C947 A0A0P8YEQ8 B4R0A9 B4I4Y3 B5X0K8 Q9VHY7 Q6NQV8 B3NYS8 B4PRD7 A0A0B4LGT2 A0A0R1E1C4 A0A0R1E1Q9 A0A1W4VYY7 A0A1W4VZM1 A0A1B0CSF7 A0A1B0CSY4 A0A1L8DIW8 A0A1B0GQ56 A0A1L8DIP3 A0A1J1IHM1 A0A1J1IG66 B0WRI4 A0A1Q3F007 A0A182QEN6 A0A182JF65 A0A182N8V8 Q7Q0M1 A0A084W5L3 A0A2M4ACK1 A0A182F493 A0A2M4ACA0 A0A2M4CRT3 A0A2M4CRM4 W5JSU2 A0A1S4H4F7 A0A182IYC8 A0A182FIN2 A0A182N0Q4 A0A084VFU2 A0A182PBH5 A0A182R283 A0A182YF07 A0A182W659 Q7QF89 A0A182MF93 A0A182XRE9 A0A182VHX2 A0A182IGN3 A0A182KA14 A0A182S924
A0A2W1BN88 A0A194QDF8 A0A0L7KRS9 A0A0L0CKM1 A0A0A1WRK9 A0A0A1WL94 A0A1A9WFM6 A0A1B0FNB9 A0A1A9YI35 A0A1A9Z6J3 B4K7I8 W8BFL5 A0A034W680 A0A0Q9XA97 A0A0K8VD15 A0A034W4P9 A0A1B0GQ57 A0A1B0BY82 A0A1I8MKT5 A0A3B0KG84 A0A3B0K7L8 A0A1I8PRH0 A0A1I8MKS7 A0A0R3NJS7 B4GMX9 A0A1I8PRL8 A0A1I8MKT1 Q294L4 A0A3B0JSR8 A0A3B0JUI7 A0A0R3NJU9 A0A0R3NJP8 I5APS4 A0A1I8PRL4 A0A336LYY4 A0A0M4EH70 B4N8D0 A0A1B0GJI4 B4JI37 B4M0V1 A0A0Q9WUI7 A0A0Q9WSR2 A0A0Q9WIQ4 B3LXF7 A0A0P9C947 A0A0P8YEQ8 B4R0A9 B4I4Y3 B5X0K8 Q9VHY7 Q6NQV8 B3NYS8 B4PRD7 A0A0B4LGT2 A0A0R1E1C4 A0A0R1E1Q9 A0A1W4VYY7 A0A1W4VZM1 A0A1B0CSF7 A0A1B0CSY4 A0A1L8DIW8 A0A1B0GQ56 A0A1L8DIP3 A0A1J1IHM1 A0A1J1IG66 B0WRI4 A0A1Q3F007 A0A182QEN6 A0A182JF65 A0A182N8V8 Q7Q0M1 A0A084W5L3 A0A2M4ACK1 A0A182F493 A0A2M4ACA0 A0A2M4CRT3 A0A2M4CRM4 W5JSU2 A0A1S4H4F7 A0A182IYC8 A0A182FIN2 A0A182N0Q4 A0A084VFU2 A0A182PBH5 A0A182R283 A0A182YF07 A0A182W659 Q7QF89 A0A182MF93 A0A182XRE9 A0A182VHX2 A0A182IGN3 A0A182KA14 A0A182S924
PDB
6MOL
E-value=1.04475e-38,
Score=405
Ontologies
GO
Topology
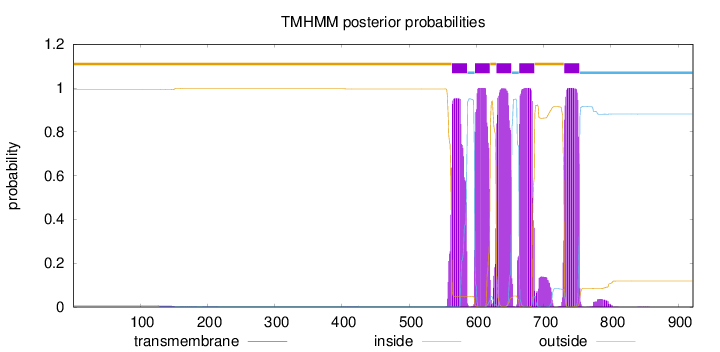
Length:
922
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.19625
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00745
outside
1 - 563
TMhelix
564 - 586
inside
587 - 597
TMhelix
598 - 620
outside
621 - 629
TMhelix
630 - 652
inside
653 - 663
TMhelix
664 - 686
outside
687 - 730
TMhelix
731 - 753
inside
754 - 922
Population Genetic Test Statistics
Pi
191.028109
Theta
178.642197
Tajima's D
-1.368705
CLR
1.540645
CSRT
0.0793460326983651
Interpretation
Uncertain
