Gene
KWMTBOMO03748 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014211
Annotation
transketolase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.959
Sequence
CDS
ATGAAGGGAGATAAAAATGTTGACTTTGAACAACTAAAAGATATCGCCAACAAACTAAGAATCGACAGTATTGTTGCTACAAACGCCTCAAAGTCTGGTCACCCGACATCATGTGCGTCAATGGCGGAGATAATGTCTGTTCTCTTCTTCCACACAATGCGGTACAAGATCTCCGCGCCACGAGACGCTTCCGCCGACAGATTTATTCTTTCCAAGGGTCACGCAGCGCCGATCCTGTACGCGGCGTGGGCGGAGGCCGGGCTGTTCCCGCTGGACGAGCTGAAGAACTTGCGCAAGCTGGACTCGGACCTGGAGGGCCACCCCACCCCGCGACTCAACTTCGTGGACGTCGGCACCGGCTCCCTGGGCCAGGGGCTCGCCGTGGCGGCCGGCATGGCCTACGTCGGGAAGTACTTCGACCAGGCGCCCTACAGGGTGTATTGCCTGGTGGGCGACGGAGAGGCAGCCGAGGGCAGCATCTGGGAGTCGCTGCACTTTGCCAGCCACTACAAGCTGGACAACCTGGTCGTCATCTTCGATGTTAACCGTCTGGGGCAGTCCGAGCCTACCTCTCTGCAACATCAGCTGGAAGTGTACGACGCGCGCCTGAAGGCGTTCGGTCTCAACTCGCTGGTGGTGGACGGACATGACGTCACGGAGCTGGTCAAGGCGTTCGACGAGGCCGCTTCAGTGACCGGGAAGCCAACAGCGCTGGTGGCTAAGACTTACAAGGGAAGGGGCTTCCCGGGGATCGAGGACCTGGACAACTGGCACGGAAAGGCTCTGGGAGCTGAGGGAGAGAAGATCATCAAGCATCTTCAGTCTCAGATCAAAACAACAAAGATTACACTAAAACCGAAAGCTCCACTAGCAGAAGCTCCCAAAGTTCACATCGATGACATCACACTCTCGTCTCCGCCAGCTTACAAGCAAGGAGAGCTGGTAGCCACAAGACTGGCATATGGAACTGCACTTAAAAAGATTGCTGATACTAATCTGCGTGTTATCGCTTTGGATGGTGACACCAAGAACTCTACATTCAGTGACAAGCTTCGGAATGCTTACCCTGAAAGATATGTGGAGTGTTTCATCGCTGAACAGAATCTAGTTGGAGTGGCTACGGGAGCAGCCTGCAGGGACCGGGCCGTGGTGTTTGTCTCCACATTTGCTGCTTTCTTCACAAGAACTTTTGACCAAATTCGTATGGGTGCTATTAGTCAGTCCAACATTAACCTTGCCGGTTCACACTGCGGAGTCAGCATCGGAGAGGACGGTCCGAGTCAGATGGGACTGGAGGACCTTGCCATGTTCCGTACAGTGCCTACTGCTACTGTCTTCTACCCGTCTGATGCAGTCTCTACAGAGCGTGCAGTAGAATTAGCAGCCAACACTCGAGGCATCTGTTACATCAGAACTTCTAGACCCAACACGGCTATCTTGTACCCCAATGATGAAGTATTCAAGGTCGGACAGGCCAAAGTACTCCGTCAGTCCGCAAAGGACCGAGTGTTGTTGATCGGAGCCGGAATCACGCTGCACGAAGCACTTGCTGCTGCTGAAGAATTGAAAAAAGAAGGTGTGGAGGCTCGTGTGTTGGACCCGTTCACCATCAAGCCATTGGACGAGGCTGCAGTTCTGGAGAATGCTCGGGCTGCTGAGGGAAGAATACTTGTTGTTGAAGATCATTATCAAGCGGGAGGTATCGGCGAGGCGGTGCTGTCGGCGGTGTCGCTGCAGCGTGACGTCATCGTGCGGCATCTGTACGTGCGCGAGGTGCCCCGCAGCGGCCCGCCCGCCGCGCTGCTCGACCACTACGGCATCTCGCAACAACACATCGTGGCCGCCGCCAAGCAACTCATTAAAGCCTAA
Protein
MKGDKNVDFEQLKDIANKLRIDSIVATNASKSGHPTSCASMAEIMSVLFFHTMRYKISAPRDASADRFILSKGHAAPILYAAWAEAGLFPLDELKNLRKLDSDLEGHPTPRLNFVDVGTGSLGQGLAVAAGMAYVGKYFDQAPYRVYCLVGDGEAAEGSIWESLHFASHYKLDNLVVIFDVNRLGQSEPTSLQHQLEVYDARLKAFGLNSLVVDGHDVTELVKAFDEAASVTGKPTALVAKTYKGRGFPGIEDLDNWHGKALGAEGEKIIKHLQSQIKTTKITLKPKAPLAEAPKVHIDDITLSSPPAYKQGELVATRLAYGTALKKIADTNLRVIALDGDTKNSTFSDKLRNAYPERYVECFIAEQNLVGVATGAACRDRAVVFVSTFAAFFTRTFDQIRMGAISQSNINLAGSHCGVSIGEDGPSQMGLEDLAMFRTVPTATVFYPSDAVSTERAVELAANTRGICYIRTSRPNTAILYPNDEVFKVGQAKVLRQSAKDRVLLIGAGITLHEALAAAEELKKEGVEARVLDPFTIKPLDEAAVLENARAAEGRILVVEDHYQAGGIGEAVLSAVSLQRDVIVRHLYVREVPRSGPPAALLDHYGISQQHIVAAAKQLIKA
Summary
Uniprot
Q2F645
A0A2W1BQN8
A0A194QJ05
A0A437BSR9
A0A212FCR2
A0A0N1I7U9
+ More
A0A1J1IPA6 S4PXN1 Q17CT0 A0A2J7QJX3 A0A2M3Z1Z5 A0A2Z5RDW8 A0A2M4A1P2 W5J901 A0A2J7QJW6 A0A2M4BH94 A0A0C9RSU3 A0A2M4BH71 A0A0C9RBP5 A0A240PMN5 W8BDC2 A0A1B0GL38 A0A1I8QD58 A0A0L0BN54 A0A1B6CRA8 A0A1A9UF47 A0A411G6I2 D3TM09 A0A067R4R5 B0WL69 A0A182Q3H8 A0A182NZM0 A0A1B0A437 A0A1B6CUX2 A0A182YB26 A0A182FKK5 A0A1Q3FEA2 A0A1Q3FEC7 A0A182N2B8 A0A182KL38 A0A310SA74 A0A1Y9J0X5 A0A182UZY7 A0A2C9GRN6 Q7QHE9 A0A1A9YRN1 T1DE26 A0A1B0B4B3 A0A1I9WLJ5 A0A0L7QLA7 A0A182TRF4 A0A151IUS6 A0A154PIP2 B4M501 A0A182M8W6 A0A1B6FLZ4 A0A182JNC0 T1PGJ4 A0A195FDH4 F4WHV1 V9IIU4 A0A2A3E7N9 A0A084VHT8 A0A195BEQ6 A0A2S2NUG9 A0A158NBZ2 A0A1I8N4E8 A0A0M8ZRQ5 B4NLA5 A0A088AUY4 A0A2H8U097 A0A3L8DV82 A0A1L8E1X3 A0A195CBH8 A0A1L8E1Z5 A0A182WDI5 A0A026VY56 J9JQV2 A0A0M5IZI9 B4JSR8 A0A2S2QGD1 A0A0A1XC73 E2C3X4 Q9VHN7 A0A0K8UPW0 B3P4V0 B4KCP2 B4PT84 B4QXZ9 A0A3B0KDM0 A0A1D2NA95 A0A1W4WRK5 A0A1W4WFF1 A0A1W4W358 B5DWE4 B3M235 K7J0A1 B4G2T4 A0A1Y1LB61 A0A232EYK6
A0A1J1IPA6 S4PXN1 Q17CT0 A0A2J7QJX3 A0A2M3Z1Z5 A0A2Z5RDW8 A0A2M4A1P2 W5J901 A0A2J7QJW6 A0A2M4BH94 A0A0C9RSU3 A0A2M4BH71 A0A0C9RBP5 A0A240PMN5 W8BDC2 A0A1B0GL38 A0A1I8QD58 A0A0L0BN54 A0A1B6CRA8 A0A1A9UF47 A0A411G6I2 D3TM09 A0A067R4R5 B0WL69 A0A182Q3H8 A0A182NZM0 A0A1B0A437 A0A1B6CUX2 A0A182YB26 A0A182FKK5 A0A1Q3FEA2 A0A1Q3FEC7 A0A182N2B8 A0A182KL38 A0A310SA74 A0A1Y9J0X5 A0A182UZY7 A0A2C9GRN6 Q7QHE9 A0A1A9YRN1 T1DE26 A0A1B0B4B3 A0A1I9WLJ5 A0A0L7QLA7 A0A182TRF4 A0A151IUS6 A0A154PIP2 B4M501 A0A182M8W6 A0A1B6FLZ4 A0A182JNC0 T1PGJ4 A0A195FDH4 F4WHV1 V9IIU4 A0A2A3E7N9 A0A084VHT8 A0A195BEQ6 A0A2S2NUG9 A0A158NBZ2 A0A1I8N4E8 A0A0M8ZRQ5 B4NLA5 A0A088AUY4 A0A2H8U097 A0A3L8DV82 A0A1L8E1X3 A0A195CBH8 A0A1L8E1Z5 A0A182WDI5 A0A026VY56 J9JQV2 A0A0M5IZI9 B4JSR8 A0A2S2QGD1 A0A0A1XC73 E2C3X4 Q9VHN7 A0A0K8UPW0 B3P4V0 B4KCP2 B4PT84 B4QXZ9 A0A3B0KDM0 A0A1D2NA95 A0A1W4WRK5 A0A1W4WFF1 A0A1W4W358 B5DWE4 B3M235 K7J0A1 B4G2T4 A0A1Y1LB61 A0A232EYK6
Pubmed
28756777
26354079
22118469
23622113
17510324
26760975
+ More
20920257 23761445 24495485 26108605 20353571 24845553 25244985 20966253 12364791 14747013 17210077 24330624 27538518 17994087 21719571 24438588 21347285 25315136 30249741 24508170 25830018 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 15632085 20075255 28004739 28648823
20920257 23761445 24495485 26108605 20353571 24845553 25244985 20966253 12364791 14747013 17210077 24330624 27538518 17994087 21719571 24438588 21347285 25315136 30249741 24508170 25830018 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 27289101 15632085 20075255 28004739 28648823
EMBL
DQ311227
ABD36172.1
KZ149996
PZC75427.1
KQ459167
KPJ03436.1
+ More
RSAL01000013 RVE53420.1 AGBW02009154 OWR51536.1 KQ460938 KPJ10677.1 CVRI01000057 CRL02067.1 GAIX01004748 JAA87812.1 CH477304 EAT44192.1 NEVH01013548 PNF28878.1 GGFM01001785 MBW22536.1 FX983731 BAX07230.1 GGFK01001386 MBW34707.1 ADMH02002090 ETN59360.1 PNF28877.1 GGFJ01003266 MBW52407.1 GBYB01010561 JAG80328.1 GGFJ01003265 MBW52406.1 GBYB01010562 JAG80329.1 GAMC01007260 JAB99295.1 AJWK01034141 JRES01001605 KNC21540.1 GEDC01021453 JAS15845.1 MH365691 QBB01500.1 EZ422461 ADD18737.1 KK852702 KDR18110.1 DS231980 EDS30242.1 AXCN02001729 GEDC01020012 JAS17286.1 GFDL01009160 JAV25885.1 GFDL01009193 JAV25852.1 KQ767066 OAD53501.1 APCN01001484 AAAB01008816 EAA05148.4 GALA01001227 JAA93625.1 JXJN01008245 KU932379 APA34015.1 KQ414934 KOC59316.1 KQ980949 KYN11221.1 KQ434926 KZC11719.1 CH940652 EDW59712.1 AXCM01001359 GECZ01027893 GECZ01018569 GECZ01016265 JAS41876.1 JAS51200.1 JAS53504.1 KA646978 AFP61607.1 KQ981636 KYN38710.1 GL888170 EGI66180.1 JR047436 AEY60416.1 KZ288356 PBC27199.1 ATLV01013235 KE524847 KFB37532.1 KQ976500 KYM83076.1 GGMR01008232 MBY20851.1 ADTU01011502 KQ435883 KOX69770.1 CH964272 EDW84308.1 GFXV01007784 MBW19589.1 QOIP01000004 RLU24123.1 GFDF01001377 JAV12707.1 KQ978068 KYM97468.1 GFDF01001343 JAV12741.1 KK107785 EZA47799.1 ABLF02039104 CP012526 ALC45644.1 CH916373 EDV94808.1 GGMS01007616 MBY76819.1 GBXI01006234 JAD08058.1 GL452364 EFN77360.1 AE014297 BT016067 AAF54265.3 AAV36952.1 GDHF01023929 JAI28385.1 CH954181 EDV49894.1 CH933806 EDW15891.1 CM000160 EDW96545.1 CM000364 EDX13736.1 OUUW01000007 SPP83111.1 LJIJ01000129 ODN02005.1 CM000070 EDY68135.2 CH902617 EDV43359.1 CH479179 EDW24129.1 GEZM01060802 JAV70899.1 NNAY01001613 OXU23423.1
RSAL01000013 RVE53420.1 AGBW02009154 OWR51536.1 KQ460938 KPJ10677.1 CVRI01000057 CRL02067.1 GAIX01004748 JAA87812.1 CH477304 EAT44192.1 NEVH01013548 PNF28878.1 GGFM01001785 MBW22536.1 FX983731 BAX07230.1 GGFK01001386 MBW34707.1 ADMH02002090 ETN59360.1 PNF28877.1 GGFJ01003266 MBW52407.1 GBYB01010561 JAG80328.1 GGFJ01003265 MBW52406.1 GBYB01010562 JAG80329.1 GAMC01007260 JAB99295.1 AJWK01034141 JRES01001605 KNC21540.1 GEDC01021453 JAS15845.1 MH365691 QBB01500.1 EZ422461 ADD18737.1 KK852702 KDR18110.1 DS231980 EDS30242.1 AXCN02001729 GEDC01020012 JAS17286.1 GFDL01009160 JAV25885.1 GFDL01009193 JAV25852.1 KQ767066 OAD53501.1 APCN01001484 AAAB01008816 EAA05148.4 GALA01001227 JAA93625.1 JXJN01008245 KU932379 APA34015.1 KQ414934 KOC59316.1 KQ980949 KYN11221.1 KQ434926 KZC11719.1 CH940652 EDW59712.1 AXCM01001359 GECZ01027893 GECZ01018569 GECZ01016265 JAS41876.1 JAS51200.1 JAS53504.1 KA646978 AFP61607.1 KQ981636 KYN38710.1 GL888170 EGI66180.1 JR047436 AEY60416.1 KZ288356 PBC27199.1 ATLV01013235 KE524847 KFB37532.1 KQ976500 KYM83076.1 GGMR01008232 MBY20851.1 ADTU01011502 KQ435883 KOX69770.1 CH964272 EDW84308.1 GFXV01007784 MBW19589.1 QOIP01000004 RLU24123.1 GFDF01001377 JAV12707.1 KQ978068 KYM97468.1 GFDF01001343 JAV12741.1 KK107785 EZA47799.1 ABLF02039104 CP012526 ALC45644.1 CH916373 EDV94808.1 GGMS01007616 MBY76819.1 GBXI01006234 JAD08058.1 GL452364 EFN77360.1 AE014297 BT016067 AAF54265.3 AAV36952.1 GDHF01023929 JAI28385.1 CH954181 EDV49894.1 CH933806 EDW15891.1 CM000160 EDW96545.1 CM000364 EDX13736.1 OUUW01000007 SPP83111.1 LJIJ01000129 ODN02005.1 CM000070 EDY68135.2 CH902617 EDV43359.1 CH479179 EDW24129.1 GEZM01060802 JAV70899.1 NNAY01001613 OXU23423.1
Proteomes
UP000053268
UP000283053
UP000007151
UP000053240
UP000183832
UP000008820
+ More
UP000235965 UP000000673 UP000075880 UP000092461 UP000095300 UP000037069 UP000078200 UP000027135 UP000002320 UP000075886 UP000075885 UP000092445 UP000076408 UP000069272 UP000075884 UP000075882 UP000076407 UP000075903 UP000075840 UP000007062 UP000092443 UP000092460 UP000053825 UP000075902 UP000078492 UP000076502 UP000008792 UP000075883 UP000075881 UP000078541 UP000007755 UP000242457 UP000030765 UP000078540 UP000005205 UP000095301 UP000053105 UP000007798 UP000005203 UP000279307 UP000078542 UP000075920 UP000053097 UP000007819 UP000092553 UP000001070 UP000008237 UP000000803 UP000008711 UP000009192 UP000002282 UP000000304 UP000268350 UP000094527 UP000192223 UP000192221 UP000001819 UP000007801 UP000002358 UP000008744 UP000215335
UP000235965 UP000000673 UP000075880 UP000092461 UP000095300 UP000037069 UP000078200 UP000027135 UP000002320 UP000075886 UP000075885 UP000092445 UP000076408 UP000069272 UP000075884 UP000075882 UP000076407 UP000075903 UP000075840 UP000007062 UP000092443 UP000092460 UP000053825 UP000075902 UP000078492 UP000076502 UP000008792 UP000075883 UP000075881 UP000078541 UP000007755 UP000242457 UP000030765 UP000078540 UP000005205 UP000095301 UP000053105 UP000007798 UP000005203 UP000279307 UP000078542 UP000075920 UP000053097 UP000007819 UP000092553 UP000001070 UP000008237 UP000000803 UP000008711 UP000009192 UP000002282 UP000000304 UP000268350 UP000094527 UP000192223 UP000192221 UP000001819 UP000007801 UP000002358 UP000008744 UP000215335
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
Q2F645
A0A2W1BQN8
A0A194QJ05
A0A437BSR9
A0A212FCR2
A0A0N1I7U9
+ More
A0A1J1IPA6 S4PXN1 Q17CT0 A0A2J7QJX3 A0A2M3Z1Z5 A0A2Z5RDW8 A0A2M4A1P2 W5J901 A0A2J7QJW6 A0A2M4BH94 A0A0C9RSU3 A0A2M4BH71 A0A0C9RBP5 A0A240PMN5 W8BDC2 A0A1B0GL38 A0A1I8QD58 A0A0L0BN54 A0A1B6CRA8 A0A1A9UF47 A0A411G6I2 D3TM09 A0A067R4R5 B0WL69 A0A182Q3H8 A0A182NZM0 A0A1B0A437 A0A1B6CUX2 A0A182YB26 A0A182FKK5 A0A1Q3FEA2 A0A1Q3FEC7 A0A182N2B8 A0A182KL38 A0A310SA74 A0A1Y9J0X5 A0A182UZY7 A0A2C9GRN6 Q7QHE9 A0A1A9YRN1 T1DE26 A0A1B0B4B3 A0A1I9WLJ5 A0A0L7QLA7 A0A182TRF4 A0A151IUS6 A0A154PIP2 B4M501 A0A182M8W6 A0A1B6FLZ4 A0A182JNC0 T1PGJ4 A0A195FDH4 F4WHV1 V9IIU4 A0A2A3E7N9 A0A084VHT8 A0A195BEQ6 A0A2S2NUG9 A0A158NBZ2 A0A1I8N4E8 A0A0M8ZRQ5 B4NLA5 A0A088AUY4 A0A2H8U097 A0A3L8DV82 A0A1L8E1X3 A0A195CBH8 A0A1L8E1Z5 A0A182WDI5 A0A026VY56 J9JQV2 A0A0M5IZI9 B4JSR8 A0A2S2QGD1 A0A0A1XC73 E2C3X4 Q9VHN7 A0A0K8UPW0 B3P4V0 B4KCP2 B4PT84 B4QXZ9 A0A3B0KDM0 A0A1D2NA95 A0A1W4WRK5 A0A1W4WFF1 A0A1W4W358 B5DWE4 B3M235 K7J0A1 B4G2T4 A0A1Y1LB61 A0A232EYK6
A0A1J1IPA6 S4PXN1 Q17CT0 A0A2J7QJX3 A0A2M3Z1Z5 A0A2Z5RDW8 A0A2M4A1P2 W5J901 A0A2J7QJW6 A0A2M4BH94 A0A0C9RSU3 A0A2M4BH71 A0A0C9RBP5 A0A240PMN5 W8BDC2 A0A1B0GL38 A0A1I8QD58 A0A0L0BN54 A0A1B6CRA8 A0A1A9UF47 A0A411G6I2 D3TM09 A0A067R4R5 B0WL69 A0A182Q3H8 A0A182NZM0 A0A1B0A437 A0A1B6CUX2 A0A182YB26 A0A182FKK5 A0A1Q3FEA2 A0A1Q3FEC7 A0A182N2B8 A0A182KL38 A0A310SA74 A0A1Y9J0X5 A0A182UZY7 A0A2C9GRN6 Q7QHE9 A0A1A9YRN1 T1DE26 A0A1B0B4B3 A0A1I9WLJ5 A0A0L7QLA7 A0A182TRF4 A0A151IUS6 A0A154PIP2 B4M501 A0A182M8W6 A0A1B6FLZ4 A0A182JNC0 T1PGJ4 A0A195FDH4 F4WHV1 V9IIU4 A0A2A3E7N9 A0A084VHT8 A0A195BEQ6 A0A2S2NUG9 A0A158NBZ2 A0A1I8N4E8 A0A0M8ZRQ5 B4NLA5 A0A088AUY4 A0A2H8U097 A0A3L8DV82 A0A1L8E1X3 A0A195CBH8 A0A1L8E1Z5 A0A182WDI5 A0A026VY56 J9JQV2 A0A0M5IZI9 B4JSR8 A0A2S2QGD1 A0A0A1XC73 E2C3X4 Q9VHN7 A0A0K8UPW0 B3P4V0 B4KCP2 B4PT84 B4QXZ9 A0A3B0KDM0 A0A1D2NA95 A0A1W4WRK5 A0A1W4WFF1 A0A1W4W358 B5DWE4 B3M235 K7J0A1 B4G2T4 A0A1Y1LB61 A0A232EYK6
PDB
3OOY
E-value=0,
Score=1884
Ontologies
PATHWAY
GO
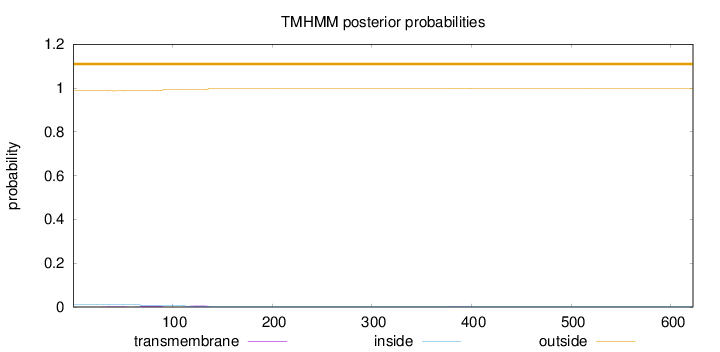
Topology
Length:
622
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.295479999999999
Exp number, first 60 AAs:
0.03953
Total prob of N-in:
0.01202
outside
1 - 622
Population Genetic Test Statistics
Pi
210.046087
Theta
165.291616
Tajima's D
0.716589
CLR
25.308561
CSRT
0.577921103944803
Interpretation
Uncertain
