Gene
KWMTBOMO03722 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA009992
Annotation
PREDICTED:_EH_domain-containing_protein_3_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.695
Sequence
CDS
ATGTTTAGTTGGCTTAAGAAAGAAGGTGAAAAAGCGGAAAATATTGAAAATGTCGTGGAAGGGCTCAAGAGGATTTACAAAACTAAACTGTTGCCTCTTGAGCTGCACTACCAGTTCCACGATTTCCATTCGCCGCAGCTCGAAGATCCAGATTTCGATGCCAAACCCATGATACTTCTCGTGGGACAGTACTCCACCGGTAAAACGACATTTATCAGGTATTTGCTAGAGAGAGATTTTCCGGGCATCCGTATTGGTCCAGAGCCGACTACAGATAGATTTATAGCCGTTATGTTCGACGAAAAAGAGGGCGTAATTCCGGGTAATGCGTTAGTAGTAGATCCGAAAAAACAGTTTCGACCATTGAGTAAATTTGGTAACGCCTTCCTGAACCGATTCCAGTGTTCAACTGTCACGTCGGACGTACTACGCGGGATCTCTATCGTTGACACACCCGGTATCCTATCTGGAGAGAAACAGCGTGTAGATCGCGGGTATGACTTTACTGGAGTATTGGAATGGTTTGCTGAGCGTGTTGACCGAATCATACTTTTGTTCGATGCTCACAAACTTGATATATCAGATGAATTTAGGCGCAGTATAGAGGCATTGAGAGGACATGATGATAAGATCCGAATTGTGCTCAACAAAGCTGATATGATAGATCATCAGCAGTTGATGCGTGTTTATGGAGCTCTGATGTGGTCACTGGGAAAAGTTTTACAAACTCCTGAGGTTGCTAGAGTGTACATTGGCTCATTTTGGGATGAGCCTCTCCGGTACGATGTTAATCGTCGACTTTTTGAAGATGAAGAACAAGATTTATTTAAAGACATGCAGTCTTTGCCAAGGAATGCTGCCCTGCGTAAACTAAACGACCTTATTAAAAGAGCACGACTTGCTAAGGTTCATGCATACATAGTCAATGAGTTAAGGAAAGAGATGCCTTCAATGTTTGGTAAAGATTCAAAAAAGAAAGATCTAATCAAGAATCTTGGTCAGGTTTATGATAAAATACAAAGAGAGCAGCAAATATCACCGGGAGACTTTCCAGATATTAAAAAAATGCAAGAGACCCTGGCCAATCATGACTTCACCAAGTTTCACCCCTTGAAACCCAAGTTGCTAGAAGTAGTGGACCACATGCTGGCAACGGACATTGCACGGCTCATGGATATGATTCCACAGGAGGACATCAACATTGTCAGTGAACCTCTGATAAAGGGTGGTGCTTTCGAAGGTGTAGAAGACCAGGTCTCACCATTCGGTTACGGTCGGGGCGAGGGCGTGGACGCCGGCCACGGTGACCCGGAATGGATCTGCAGCAAGGAGAAACCGAACTACGACCGAATCTTCCAATCTCTGAACCCGATCGACGGCAAAGTGACTGGCGCCGCCGCAAAGACAGAGATGGTGAAGTCGAAGCTGCCCAACTCTGTGCTCGGCAAGATTTGGAAGCTCTCCGACATCGACAAGGACGGCTTCCTGGACGAAGAGGAGTTTGCGCTCGCGATGCATCTCATCTCGGTGAAGGTGGACGGACACGACCTGCCCACCGAGCTGCCCTCGCACCTCGTGCCGCCCTCCAAACGGAACTGA
Protein
MFSWLKKEGEKAENIENVVEGLKRIYKTKLLPLELHYQFHDFHSPQLEDPDFDAKPMILLVGQYSTGKTTFIRYLLERDFPGIRIGPEPTTDRFIAVMFDEKEGVIPGNALVVDPKKQFRPLSKFGNAFLNRFQCSTVTSDVLRGISIVDTPGILSGEKQRVDRGYDFTGVLEWFAERVDRIILLFDAHKLDISDEFRRSIEALRGHDDKIRIVLNKADMIDHQQLMRVYGALMWSLGKVLQTPEVARVYIGSFWDEPLRYDVNRRLFEDEEQDLFKDMQSLPRNAALRKLNDLIKRARLAKVHAYIVNELRKEMPSMFGKDSKKKDLIKNLGQVYDKIQREQQISPGDFPDIKKMQETLANHDFTKFHPLKPKLLEVVDHMLATDIARLMDMIPQEDINIVSEPLIKGGAFEGVEDQVSPFGYGRGEGVDAGHGDPEWICSKEKPNYDRIFQSLNPIDGKVTGAAAKTEMVKSKLPNSVLGKIWKLSDIDKDGFLDEEEFALAMHLISVKVDGHDLPTELPSHLVPPSKRN
Summary
Similarity
Belongs to the TRAFAC class dynamin-like GTPase superfamily. Dynamin/Fzo/YdjA family.
Belongs to the histidine acid phosphatase family.
Belongs to the histidine acid phosphatase family.
Uniprot
A0A194QEQ2
A0A220K8I4
A0A3S2NZX2
E2AZE9
A0A151WJF0
A0A0J7N331
+ More
A0A195CQM0 A0A2J7R596 E2C8F3 A0A026WV74 F4WGA5 A0A195B693 A0A158NI61 A0A232EL11 E9IB79 A0A195EGP8 A0A151JXC0 Q16T24 A0A1W4WR69 A0A023ETP5 A0A154P3I4 A0A1J1IJV4 U5EMK8 A0A2A3EAJ6 V9IMV7 A0A088A395 A0A1Q3FC31 B0WNC3 A0A411G820 A0A1Y1N381 A0A0T6AXK7 A0A084W7R4 A0A1Q3FC63 A0A0M9A7H5 K7J964 B4KC47 A0A310SEK6 Q7Q7M8 A0A1Y9GKD8 A0A182VJ75 A0A182LNL6 R4G5H8 Q8T6I0 A0A182Q189 Q8IGN0 B4MBB5 B4NI15 Q299V4 A0A034W1B4 A0A0L7QXR0 R4WJU9 B3M1W0 A0A0L0CSV7 B4QS70 B4HGF8 B3P472 Q8T8W3 B4PPE1 A0A2M4CTJ3 A0A2M3Z4V5 A0A336K5Q6 T1P861 A0A1L8ECI4 A0A1B0CRC7 A0A1I8PMV3 A0A139WP92 A0A1L8ECY2 A0A2M4AEW8 A0A3B0K9Z4 A0A1W4UMX8 A0A1L8DM80 A0A0K8VR18 A0A1S4GM49 E0VWS6 A0A0A1XQJ0 W8C0Q9 A0A0A9YYD3 A0A0P4VH63 D3TMZ1 A0A1A9VS50 A0A1A9ZGE4 A0A0K8TAD0 A0A1A9XDL2 A0A0A9Z0A3 A0A2M4AFA1 A0A0A9W3E3 A0A0P6I8F2 A0A0P5B0F6 A0A0K8SCA0 E9GJ29 N6T8R1 A0A0A9YXZ4 A0A0M4ECQ3 A0A0P5ZNQ1 A0A023FLZ2 V5HPU0 A0A0P6BUH4 A0A023GL46 A0A1E1X9N6 A0A131Y206
A0A195CQM0 A0A2J7R596 E2C8F3 A0A026WV74 F4WGA5 A0A195B693 A0A158NI61 A0A232EL11 E9IB79 A0A195EGP8 A0A151JXC0 Q16T24 A0A1W4WR69 A0A023ETP5 A0A154P3I4 A0A1J1IJV4 U5EMK8 A0A2A3EAJ6 V9IMV7 A0A088A395 A0A1Q3FC31 B0WNC3 A0A411G820 A0A1Y1N381 A0A0T6AXK7 A0A084W7R4 A0A1Q3FC63 A0A0M9A7H5 K7J964 B4KC47 A0A310SEK6 Q7Q7M8 A0A1Y9GKD8 A0A182VJ75 A0A182LNL6 R4G5H8 Q8T6I0 A0A182Q189 Q8IGN0 B4MBB5 B4NI15 Q299V4 A0A034W1B4 A0A0L7QXR0 R4WJU9 B3M1W0 A0A0L0CSV7 B4QS70 B4HGF8 B3P472 Q8T8W3 B4PPE1 A0A2M4CTJ3 A0A2M3Z4V5 A0A336K5Q6 T1P861 A0A1L8ECI4 A0A1B0CRC7 A0A1I8PMV3 A0A139WP92 A0A1L8ECY2 A0A2M4AEW8 A0A3B0K9Z4 A0A1W4UMX8 A0A1L8DM80 A0A0K8VR18 A0A1S4GM49 E0VWS6 A0A0A1XQJ0 W8C0Q9 A0A0A9YYD3 A0A0P4VH63 D3TMZ1 A0A1A9VS50 A0A1A9ZGE4 A0A0K8TAD0 A0A1A9XDL2 A0A0A9Z0A3 A0A2M4AFA1 A0A0A9W3E3 A0A0P6I8F2 A0A0P5B0F6 A0A0K8SCA0 E9GJ29 N6T8R1 A0A0A9YXZ4 A0A0M4ECQ3 A0A0P5ZNQ1 A0A023FLZ2 V5HPU0 A0A0P6BUH4 A0A023GL46 A0A1E1X9N6 A0A131Y206
Pubmed
26354079
28630352
20798317
24508170
21719571
21347285
+ More
28648823 21282665 17510324 24945155 28004739 24438588 20075255 17994087 12364791 20966253 10731132 12537568 12537572 12537573 12537574 15155807 16110336 17569856 17569867 26109357 26109356 15632085 25348373 23691247 26108605 17550304 25315136 18362917 19820115 20566863 25830018 24495485 25401762 27129103 20353571 26823975 21292972 23537049 25765539 28503490
28648823 21282665 17510324 24945155 28004739 24438588 20075255 17994087 12364791 20966253 10731132 12537568 12537572 12537573 12537574 15155807 16110336 17569856 17569867 26109357 26109356 15632085 25348373 23691247 26108605 17550304 25315136 18362917 19820115 20566863 25830018 24495485 25401762 27129103 20353571 26823975 21292972 23537049 25765539 28503490
EMBL
KQ459167
KPJ03425.1
MF319687
ASJ26401.1
RSAL01000013
RVE53411.1
+ More
GL444207 EFN61188.1 KQ983039 KYQ47982.1 LBMM01011006 KMQ87085.1 KQ977481 KYN02409.1 NEVH01007392 PNF36009.1 GL453666 EFN75788.1 KK107111 EZA59004.1 GL888128 EGI66872.1 KQ976579 KYM80026.1 ADTU01016304 NNAY01003689 OXU19002.1 GL762111 EFZ22199.1 KQ978957 KYN27059.1 KQ981578 KYN39752.1 CH477661 EAT37607.1 GAPW01000883 JAC12715.1 KQ434809 KZC06431.1 CVRI01000054 CRL00034.1 GANO01000984 JAB58887.1 KZ288322 PBC28216.1 JR053683 AEY61994.1 GFDL01009904 JAV25141.1 DS232010 EDS31577.1 MH366186 QBB01995.1 GEZM01013851 GEZM01013850 JAV92289.1 LJIG01022673 KRT79319.1 ATLV01021281 ATLV01021282 ATLV01021283 ATLV01021284 KE525316 KFB46258.1 GFDL01009936 JAV25109.1 KQ435719 KOX78810.1 AAZX01002217 AAZX01010953 AAZX01014300 CH933806 EDW16920.2 KQ770178 OAD52669.1 AAAB01008952 EAA10568.5 APCN01005960 APCN01005961 ACPB03000063 GAHY01000560 JAA76950.1 AE014297 AF473822 AAF54856.2 AAL85325.1 AXCN02002206 BT001695 AAN71450.1 CH940656 EDW58386.2 CH964272 EDW83665.1 CM000070 EAL27597.4 GAKP01010480 GAKP01010478 JAC48474.1 KQ414704 KOC63336.1 AK417885 BAN21100.1 CH902617 EDV42220.2 JRES01000062 KNC34489.1 CM000364 EDX13166.1 CH480815 EDW42406.1 CH954181 EDV49317.1 AY075248 AAL68115.1 AAN13552.1 CM000160 EDW97147.1 GGFL01004407 MBW68585.1 GGFM01002737 MBW23488.1 UFQS01000109 UFQT01000109 SSW99771.1 SSX20151.1 KA644851 AFP59480.1 GFDG01002368 JAV16431.1 AJWK01024616 AJWK01024617 AJWK01024618 KQ971307 KYB29743.1 GFDG01002366 JAV16433.1 GGFK01006003 MBW39324.1 OUUW01000005 SPP80378.1 GFDF01006599 JAV07485.1 GDHF01033411 GDHF01011284 JAI18903.1 JAI41030.1 DS235824 EEB17832.1 GBXI01003934 GBXI01000693 JAD10358.1 JAD13599.1 GAMC01011347 GAMC01011346 GAMC01011345 JAB95209.1 GBHO01006998 GBHO01000480 JAG36606.1 JAG43124.1 GDKW01003733 JAI52862.1 EZ422793 ADD19069.1 GBRD01015427 GBRD01003333 JAG62488.1 GBHO01041262 GBHO01005705 GBRD01003337 GBRD01003336 GDHC01016107 GDHC01006801 JAG02342.1 JAG37899.1 JAG62484.1 JAQ02522.1 JAQ11828.1 GGFK01005997 MBW39318.1 GBHO01041265 GBHO01041260 JAG02339.1 JAG02344.1 GDIQ01008070 JAN86667.1 GDIP01206076 GDIQ01192614 GDIQ01120692 LRGB01001253 JAJ17326.1 JAK59111.1 KZS13220.1 GBRD01015426 GBRD01003335 JAG50400.1 GL732547 EFX80375.1 APGK01047050 KB741077 KB632384 ENN74083.1 ERL94214.1 GBHO01006995 GBHO01005712 GBRD01003338 GBRD01001426 JAG36609.1 JAG37892.1 JAG62483.1 CP012526 ALC45466.1 GDIP01041789 JAM61926.1 GBBK01002293 JAC22189.1 GANP01006706 JAB77762.1 GDIP01014121 JAM89594.1 GBBM01000812 JAC34606.1 GFAC01003220 JAT95968.1 GEFM01002879 JAP72917.1
GL444207 EFN61188.1 KQ983039 KYQ47982.1 LBMM01011006 KMQ87085.1 KQ977481 KYN02409.1 NEVH01007392 PNF36009.1 GL453666 EFN75788.1 KK107111 EZA59004.1 GL888128 EGI66872.1 KQ976579 KYM80026.1 ADTU01016304 NNAY01003689 OXU19002.1 GL762111 EFZ22199.1 KQ978957 KYN27059.1 KQ981578 KYN39752.1 CH477661 EAT37607.1 GAPW01000883 JAC12715.1 KQ434809 KZC06431.1 CVRI01000054 CRL00034.1 GANO01000984 JAB58887.1 KZ288322 PBC28216.1 JR053683 AEY61994.1 GFDL01009904 JAV25141.1 DS232010 EDS31577.1 MH366186 QBB01995.1 GEZM01013851 GEZM01013850 JAV92289.1 LJIG01022673 KRT79319.1 ATLV01021281 ATLV01021282 ATLV01021283 ATLV01021284 KE525316 KFB46258.1 GFDL01009936 JAV25109.1 KQ435719 KOX78810.1 AAZX01002217 AAZX01010953 AAZX01014300 CH933806 EDW16920.2 KQ770178 OAD52669.1 AAAB01008952 EAA10568.5 APCN01005960 APCN01005961 ACPB03000063 GAHY01000560 JAA76950.1 AE014297 AF473822 AAF54856.2 AAL85325.1 AXCN02002206 BT001695 AAN71450.1 CH940656 EDW58386.2 CH964272 EDW83665.1 CM000070 EAL27597.4 GAKP01010480 GAKP01010478 JAC48474.1 KQ414704 KOC63336.1 AK417885 BAN21100.1 CH902617 EDV42220.2 JRES01000062 KNC34489.1 CM000364 EDX13166.1 CH480815 EDW42406.1 CH954181 EDV49317.1 AY075248 AAL68115.1 AAN13552.1 CM000160 EDW97147.1 GGFL01004407 MBW68585.1 GGFM01002737 MBW23488.1 UFQS01000109 UFQT01000109 SSW99771.1 SSX20151.1 KA644851 AFP59480.1 GFDG01002368 JAV16431.1 AJWK01024616 AJWK01024617 AJWK01024618 KQ971307 KYB29743.1 GFDG01002366 JAV16433.1 GGFK01006003 MBW39324.1 OUUW01000005 SPP80378.1 GFDF01006599 JAV07485.1 GDHF01033411 GDHF01011284 JAI18903.1 JAI41030.1 DS235824 EEB17832.1 GBXI01003934 GBXI01000693 JAD10358.1 JAD13599.1 GAMC01011347 GAMC01011346 GAMC01011345 JAB95209.1 GBHO01006998 GBHO01000480 JAG36606.1 JAG43124.1 GDKW01003733 JAI52862.1 EZ422793 ADD19069.1 GBRD01015427 GBRD01003333 JAG62488.1 GBHO01041262 GBHO01005705 GBRD01003337 GBRD01003336 GDHC01016107 GDHC01006801 JAG02342.1 JAG37899.1 JAG62484.1 JAQ02522.1 JAQ11828.1 GGFK01005997 MBW39318.1 GBHO01041265 GBHO01041260 JAG02339.1 JAG02344.1 GDIQ01008070 JAN86667.1 GDIP01206076 GDIQ01192614 GDIQ01120692 LRGB01001253 JAJ17326.1 JAK59111.1 KZS13220.1 GBRD01015426 GBRD01003335 JAG50400.1 GL732547 EFX80375.1 APGK01047050 KB741077 KB632384 ENN74083.1 ERL94214.1 GBHO01006995 GBHO01005712 GBRD01003338 GBRD01001426 JAG36609.1 JAG37892.1 JAG62483.1 CP012526 ALC45466.1 GDIP01041789 JAM61926.1 GBBK01002293 JAC22189.1 GANP01006706 JAB77762.1 GDIP01014121 JAM89594.1 GBBM01000812 JAC34606.1 GFAC01003220 JAT95968.1 GEFM01002879 JAP72917.1
Proteomes
UP000053268
UP000283053
UP000000311
UP000075809
UP000036403
UP000078542
+ More
UP000235965 UP000008237 UP000053097 UP000007755 UP000078540 UP000005205 UP000215335 UP000078492 UP000078541 UP000008820 UP000192223 UP000076502 UP000183832 UP000242457 UP000005203 UP000002320 UP000030765 UP000053105 UP000002358 UP000009192 UP000007062 UP000075840 UP000075903 UP000075882 UP000015103 UP000000803 UP000075886 UP000008792 UP000007798 UP000001819 UP000053825 UP000007801 UP000037069 UP000000304 UP000001292 UP000008711 UP000002282 UP000095301 UP000092461 UP000095300 UP000007266 UP000268350 UP000192221 UP000009046 UP000078200 UP000092445 UP000092443 UP000076858 UP000000305 UP000019118 UP000030742 UP000092553
UP000235965 UP000008237 UP000053097 UP000007755 UP000078540 UP000005205 UP000215335 UP000078492 UP000078541 UP000008820 UP000192223 UP000076502 UP000183832 UP000242457 UP000005203 UP000002320 UP000030765 UP000053105 UP000002358 UP000009192 UP000007062 UP000075840 UP000075903 UP000075882 UP000015103 UP000000803 UP000075886 UP000008792 UP000007798 UP000001819 UP000053825 UP000007801 UP000037069 UP000000304 UP000001292 UP000008711 UP000002282 UP000095301 UP000092461 UP000095300 UP000007266 UP000268350 UP000192221 UP000009046 UP000078200 UP000092445 UP000092443 UP000076858 UP000000305 UP000019118 UP000030742 UP000092553
Interpro
Gene 3D
ProteinModelPortal
A0A194QEQ2
A0A220K8I4
A0A3S2NZX2
E2AZE9
A0A151WJF0
A0A0J7N331
+ More
A0A195CQM0 A0A2J7R596 E2C8F3 A0A026WV74 F4WGA5 A0A195B693 A0A158NI61 A0A232EL11 E9IB79 A0A195EGP8 A0A151JXC0 Q16T24 A0A1W4WR69 A0A023ETP5 A0A154P3I4 A0A1J1IJV4 U5EMK8 A0A2A3EAJ6 V9IMV7 A0A088A395 A0A1Q3FC31 B0WNC3 A0A411G820 A0A1Y1N381 A0A0T6AXK7 A0A084W7R4 A0A1Q3FC63 A0A0M9A7H5 K7J964 B4KC47 A0A310SEK6 Q7Q7M8 A0A1Y9GKD8 A0A182VJ75 A0A182LNL6 R4G5H8 Q8T6I0 A0A182Q189 Q8IGN0 B4MBB5 B4NI15 Q299V4 A0A034W1B4 A0A0L7QXR0 R4WJU9 B3M1W0 A0A0L0CSV7 B4QS70 B4HGF8 B3P472 Q8T8W3 B4PPE1 A0A2M4CTJ3 A0A2M3Z4V5 A0A336K5Q6 T1P861 A0A1L8ECI4 A0A1B0CRC7 A0A1I8PMV3 A0A139WP92 A0A1L8ECY2 A0A2M4AEW8 A0A3B0K9Z4 A0A1W4UMX8 A0A1L8DM80 A0A0K8VR18 A0A1S4GM49 E0VWS6 A0A0A1XQJ0 W8C0Q9 A0A0A9YYD3 A0A0P4VH63 D3TMZ1 A0A1A9VS50 A0A1A9ZGE4 A0A0K8TAD0 A0A1A9XDL2 A0A0A9Z0A3 A0A2M4AFA1 A0A0A9W3E3 A0A0P6I8F2 A0A0P5B0F6 A0A0K8SCA0 E9GJ29 N6T8R1 A0A0A9YXZ4 A0A0M4ECQ3 A0A0P5ZNQ1 A0A023FLZ2 V5HPU0 A0A0P6BUH4 A0A023GL46 A0A1E1X9N6 A0A131Y206
A0A195CQM0 A0A2J7R596 E2C8F3 A0A026WV74 F4WGA5 A0A195B693 A0A158NI61 A0A232EL11 E9IB79 A0A195EGP8 A0A151JXC0 Q16T24 A0A1W4WR69 A0A023ETP5 A0A154P3I4 A0A1J1IJV4 U5EMK8 A0A2A3EAJ6 V9IMV7 A0A088A395 A0A1Q3FC31 B0WNC3 A0A411G820 A0A1Y1N381 A0A0T6AXK7 A0A084W7R4 A0A1Q3FC63 A0A0M9A7H5 K7J964 B4KC47 A0A310SEK6 Q7Q7M8 A0A1Y9GKD8 A0A182VJ75 A0A182LNL6 R4G5H8 Q8T6I0 A0A182Q189 Q8IGN0 B4MBB5 B4NI15 Q299V4 A0A034W1B4 A0A0L7QXR0 R4WJU9 B3M1W0 A0A0L0CSV7 B4QS70 B4HGF8 B3P472 Q8T8W3 B4PPE1 A0A2M4CTJ3 A0A2M3Z4V5 A0A336K5Q6 T1P861 A0A1L8ECI4 A0A1B0CRC7 A0A1I8PMV3 A0A139WP92 A0A1L8ECY2 A0A2M4AEW8 A0A3B0K9Z4 A0A1W4UMX8 A0A1L8DM80 A0A0K8VR18 A0A1S4GM49 E0VWS6 A0A0A1XQJ0 W8C0Q9 A0A0A9YYD3 A0A0P4VH63 D3TMZ1 A0A1A9VS50 A0A1A9ZGE4 A0A0K8TAD0 A0A1A9XDL2 A0A0A9Z0A3 A0A2M4AFA1 A0A0A9W3E3 A0A0P6I8F2 A0A0P5B0F6 A0A0K8SCA0 E9GJ29 N6T8R1 A0A0A9YXZ4 A0A0M4ECQ3 A0A0P5ZNQ1 A0A023FLZ2 V5HPU0 A0A0P6BUH4 A0A023GL46 A0A1E1X9N6 A0A131Y206
PDB
5MVF
E-value=0,
Score=1884
Ontologies
GO
PANTHER
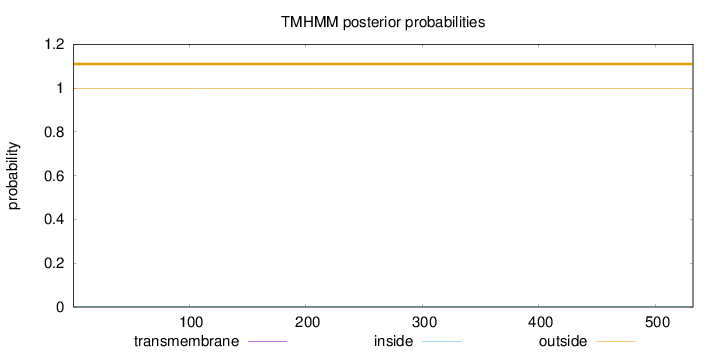
Topology
Length:
532
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00304
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.00100
outside
1 - 532
Population Genetic Test Statistics
Pi
245.817492
Theta
19.910007
Tajima's D
1.013765
CLR
0.202743
CSRT
0.674666266686666
Interpretation
Uncertain
