Gene
KWMTBOMO03721 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA010000
Annotation
PREDICTED:_small_glutamine-rich_tetratricopeptide_repeat-containing_protein_alpha-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.686 Nuclear Reliability : 1.459
Sequence
CDS
ATGTCAAACGAAATCAAAATAGTTGCAGTGAGTATCGTGAACTTTCTAAAACAGCAATTGGACGGCGATTCTTTGAGCCCTGATTCTAGAGAGAGTGTCGAAGTCGGCGTGCAATGCATTGAGACCGCTTTCGAGCTGACGACGGAAGATGCGGCTCGAGGCCTGGACTTACTGCAGCTTGTCCGTCAGCGACTGGGGCCGCCCGCTAACAAGGCTGAAGCCGAACGACTCAAGAACGAAGGCAACGAGCTCATGAAAGCTGAGCGTTACAACGAAGCGCTTGAAAAGTACACGCGCGCCATAGAGATCGACCCAAGAAACGCAGTCTACTACTGCAATCGCGCGGCTGCGCATTTCAGGCTGTGCGACCCCGAGGCAGCGGTGAGCGACTGTACCACAGCACTCGCCCTACAGCCTGACTATAGCAAGGCGTACGGACGCCTTGGGCTCGCGCTCACCGCCCTCGAGAGACACTGCGAGGCGCGCGCTGCCTACGCCCGCGCCGCTCAACTCGACCCTGAGAACGAGTCCTACCGAGAAAACCTCCAACTCGCTGACCAAACCTTAGCACAGCGCCCTAACGATCTGAGTGGCTTACTCCGGAATCCTGCTTTGATCAACATGGCTACGGAGATGCTCACTAACCCTAATATGCAGAACCTCATTTCAGGGTTGATGACAACGACAGGGGGAGCTCAGGGTGTAGGAGAGGTGCCCCCAGGAGGTATTAATGCGTTGTTAGAAGTGGGACAATCATTGGCACAACAAATGCAAGCTGCTAATCCTGATTTGGTAGAGCAACTGCGACGTCAGCTTCGACCCCCAGGGTCTGGTGAAAATCAGCCACCTTCTCAGTGA
Protein
MSNEIKIVAVSIVNFLKQQLDGDSLSPDSRESVEVGVQCIETAFELTTEDAARGLDLLQLVRQRLGPPANKAEAERLKNEGNELMKAERYNEALEKYTRAIEIDPRNAVYYCNRAAAHFRLCDPEAAVSDCTTALALQPDYSKAYGRLGLALTALERHCEARAAYARAAQLDPENESYRENLQLADQTLAQRPNDLSGLLRNPALINMATEMLTNPNMQNLISGLMTTTGGAQGVGEVPPGGINALLEVGQSLAQQMQAANPDLVEQLRRQLRPPGSGENQPPSQ
Summary
Uniprot
H9JKE9
A0A0N1IEJ1
A0A2H1WII2
A0A194QD05
I4DNY8
A0A2W1BS03
+ More
A0A195EG36 A0A0J7KVT5 F4WQS1 A0A195ESK8 A0A158NRT4 A0A151X3E4 A0A088A7V2 A0A2A3EQV3 A0A195D161 A0A026WCS8 E9IY50 A0A195BRL0 E2A9W5 A0A0N0U5G8 A0A0C9QV25 A0A0L7RBH4 A0A0P4Z5S2 A0A0N8C3W3 A0A0P5G8Y8 A0A0P5BCF9 A0A0P5KH32 A0A0P5KUX9 A0A0P5EJB2 A0A0P5PI74 A0A0P6EJJ2 A0A0P5K871 A0A0P5EQY9 A0A0P5U7G5 A0A023FVG0 A0A1E1XS65 A0A0P6G5E5 A0A0P5GWG1 A0A154PMX7 A0A0P5ZUT9 A0A2A4K2K7 A0A1E1X4T5 A0A131Z7R9 K7J8G8 A0A0P5ENI3 A0A023GKN8 G3MNU7 A0A0N8CT87 A0A0P5K7L3 A0A0P5KQN5 E3TDQ6 A0A131XMH1 A0A2R5LDG3 A0A401RGA2 A0A224Z814 B7Q815 A0A0K8RJW9 Q6NTZ8 G5BZC6 A0A0N8EUU4 Q4PLZ5 A0A0P6JC79 E3TC54 A0A293MXJ0 A0A0P6BV29 A0A3B4EHG4 A0A0P7Y7C1 W5LWG4 A0A2R2MQB0 A0A0P5JR22 H3A0N6 F7E1T8 A0A087R350 A0A0A0ABZ4 A0A286XEB3 A0A091SG92 A0A091VG92 G3WEQ9 A0A093QBY9 A0A094NEQ9 A0A091KCL6 A0A093QQ27 A0A093JCD8 K7F3S2 H0YRE4 A0A091W302 A0A452HA73 A0A091NN40 A0A091UGG0 A0A093HDL5 K7F3S1 A0A091N9R5 A0A091RP19 U3K5A4 A0A091TJV9 A0A091LU97 A0A218V971 H0YXC6 A0A093PSW1
A0A195EG36 A0A0J7KVT5 F4WQS1 A0A195ESK8 A0A158NRT4 A0A151X3E4 A0A088A7V2 A0A2A3EQV3 A0A195D161 A0A026WCS8 E9IY50 A0A195BRL0 E2A9W5 A0A0N0U5G8 A0A0C9QV25 A0A0L7RBH4 A0A0P4Z5S2 A0A0N8C3W3 A0A0P5G8Y8 A0A0P5BCF9 A0A0P5KH32 A0A0P5KUX9 A0A0P5EJB2 A0A0P5PI74 A0A0P6EJJ2 A0A0P5K871 A0A0P5EQY9 A0A0P5U7G5 A0A023FVG0 A0A1E1XS65 A0A0P6G5E5 A0A0P5GWG1 A0A154PMX7 A0A0P5ZUT9 A0A2A4K2K7 A0A1E1X4T5 A0A131Z7R9 K7J8G8 A0A0P5ENI3 A0A023GKN8 G3MNU7 A0A0N8CT87 A0A0P5K7L3 A0A0P5KQN5 E3TDQ6 A0A131XMH1 A0A2R5LDG3 A0A401RGA2 A0A224Z814 B7Q815 A0A0K8RJW9 Q6NTZ8 G5BZC6 A0A0N8EUU4 Q4PLZ5 A0A0P6JC79 E3TC54 A0A293MXJ0 A0A0P6BV29 A0A3B4EHG4 A0A0P7Y7C1 W5LWG4 A0A2R2MQB0 A0A0P5JR22 H3A0N6 F7E1T8 A0A087R350 A0A0A0ABZ4 A0A286XEB3 A0A091SG92 A0A091VG92 G3WEQ9 A0A093QBY9 A0A094NEQ9 A0A091KCL6 A0A093QQ27 A0A093JCD8 K7F3S2 H0YRE4 A0A091W302 A0A452HA73 A0A091NN40 A0A091UGG0 A0A093HDL5 K7F3S1 A0A091N9R5 A0A091RP19 U3K5A4 A0A091TJV9 A0A091LU97 A0A218V971 H0YXC6 A0A093PSW1
Pubmed
EMBL
BABH01040901
BABH01040902
KQ460938
KPJ10689.1
ODYU01008897
SOQ52883.1
+ More
KQ459167 KPJ03423.1 AK403202 BAM19628.1 KZ149996 PZC75436.1 KQ978983 KYN26837.1 LBMM01002618 KMQ94582.1 GL888275 EGI63434.1 KQ981993 KYN30892.1 ADTU01024304 KQ982562 KYQ54935.1 KZ288203 PBC33421.1 KQ977041 KYN06139.1 KK107274 QOIP01000008 EZA53743.1 RLU19711.1 GL766908 EFZ14489.1 KQ976424 KYM88581.1 GL437965 EFN69774.1 KQ435793 KOX74174.1 GBYB01004512 JAG74279.1 KQ414617 KOC68188.1 GDIP01221901 JAJ01501.1 GDIQ01117355 LRGB01000725 JAL34371.1 KZS16258.1 GDIQ01266128 GDIQ01131498 GDIQ01121794 JAJ85596.1 GDIP01191748 JAJ31654.1 GDIQ01184224 JAK67501.1 GDIQ01184225 JAK67500.1 GDIQ01269496 JAJ82228.1 GDIQ01138239 JAL13487.1 GDIQ01062767 JAN31970.1 GDIQ01193368 JAK58357.1 GDIQ01265792 JAJ85932.1 GDIP01118554 JAL85160.1 GBBL01001459 JAC25861.1 GFAA01001277 JAU02158.1 GDIQ01038131 JAN56606.1 GDIQ01236006 JAK15719.1 KQ434977 KZC12814.1 GDIP01039266 JAM64449.1 NWSH01000199 PCG78485.1 GFAC01004916 JAT94272.1 GEDV01002346 JAP86211.1 AAZX01004047 AAZX01004223 AAZX01005218 AAZX01011703 AAZX01012332 AAZX01015164 AAZX01021074 GDIP01145454 JAJ77948.1 GBBM01001745 JAC33673.1 JO843548 AEO35165.1 GDIP01101151 JAM02564.1 GDIQ01187977 JAK63748.1 GDIQ01187976 JAK63749.1 GU588486 ADO28442.1 GEFH01000332 JAP68249.1 GGLE01003444 MBY07570.1 BEZZ01001296 GCC17192.1 GFPF01011787 MAA22933.1 ABJB010215649 ABJB010561786 ABJB010871661 ABJB010913234 ABJB011100466 DS879898 EEC14987.1 GADI01002411 GEFM01005450 JAA71397.1 JAP70346.1 BC068804 AAH68804.1 JH172536 EHB14637.1 GEBF01000611 JAO03022.1 DQ066333 AAY66970.1 GDIQ01009398 JAN85339.1 GU587933 ADO27890.1 GFWV01020518 MAA45246.1 GDIP01017114 JAM86601.1 JARO02009485 KPP61580.1 AHAT01011293 GDIQ01195214 JAK56511.1 AFYH01105490 AFYH01105491 AFYH01105492 AFYH01105493 KL226068 KFM07904.1 KL871433 KGL92059.1 AAKN02057175 KK471328 KFQ57624.1 KL410953 KFR01448.1 AEFK01069977 AEFK01069978 KL671190 KFW81622.1 KL273096 KFZ65026.1 KK502193 KFP21496.1 KL224509 KFW60814.1 KK579672 KFW12710.1 AGCU01099544 AGCU01099545 ABQF01060646 ABQF01060647 ABQF01060648 ABQF01060649 ABQF01060650 KK734525 KFR09405.1 KL387282 KFP90344.1 KK432063 KFQ89232.1 KL205953 KFV77530.1 KK844249 KFP86176.1 KK803132 KFQ30242.1 AGTO01020873 KK454255 KFQ75397.1 KK507983 KFP62032.1 MUZQ01000025 OWK62533.1 ABQF01033289 ABQF01033290 KL669021 KFW75147.1
KQ459167 KPJ03423.1 AK403202 BAM19628.1 KZ149996 PZC75436.1 KQ978983 KYN26837.1 LBMM01002618 KMQ94582.1 GL888275 EGI63434.1 KQ981993 KYN30892.1 ADTU01024304 KQ982562 KYQ54935.1 KZ288203 PBC33421.1 KQ977041 KYN06139.1 KK107274 QOIP01000008 EZA53743.1 RLU19711.1 GL766908 EFZ14489.1 KQ976424 KYM88581.1 GL437965 EFN69774.1 KQ435793 KOX74174.1 GBYB01004512 JAG74279.1 KQ414617 KOC68188.1 GDIP01221901 JAJ01501.1 GDIQ01117355 LRGB01000725 JAL34371.1 KZS16258.1 GDIQ01266128 GDIQ01131498 GDIQ01121794 JAJ85596.1 GDIP01191748 JAJ31654.1 GDIQ01184224 JAK67501.1 GDIQ01184225 JAK67500.1 GDIQ01269496 JAJ82228.1 GDIQ01138239 JAL13487.1 GDIQ01062767 JAN31970.1 GDIQ01193368 JAK58357.1 GDIQ01265792 JAJ85932.1 GDIP01118554 JAL85160.1 GBBL01001459 JAC25861.1 GFAA01001277 JAU02158.1 GDIQ01038131 JAN56606.1 GDIQ01236006 JAK15719.1 KQ434977 KZC12814.1 GDIP01039266 JAM64449.1 NWSH01000199 PCG78485.1 GFAC01004916 JAT94272.1 GEDV01002346 JAP86211.1 AAZX01004047 AAZX01004223 AAZX01005218 AAZX01011703 AAZX01012332 AAZX01015164 AAZX01021074 GDIP01145454 JAJ77948.1 GBBM01001745 JAC33673.1 JO843548 AEO35165.1 GDIP01101151 JAM02564.1 GDIQ01187977 JAK63748.1 GDIQ01187976 JAK63749.1 GU588486 ADO28442.1 GEFH01000332 JAP68249.1 GGLE01003444 MBY07570.1 BEZZ01001296 GCC17192.1 GFPF01011787 MAA22933.1 ABJB010215649 ABJB010561786 ABJB010871661 ABJB010913234 ABJB011100466 DS879898 EEC14987.1 GADI01002411 GEFM01005450 JAA71397.1 JAP70346.1 BC068804 AAH68804.1 JH172536 EHB14637.1 GEBF01000611 JAO03022.1 DQ066333 AAY66970.1 GDIQ01009398 JAN85339.1 GU587933 ADO27890.1 GFWV01020518 MAA45246.1 GDIP01017114 JAM86601.1 JARO02009485 KPP61580.1 AHAT01011293 GDIQ01195214 JAK56511.1 AFYH01105490 AFYH01105491 AFYH01105492 AFYH01105493 KL226068 KFM07904.1 KL871433 KGL92059.1 AAKN02057175 KK471328 KFQ57624.1 KL410953 KFR01448.1 AEFK01069977 AEFK01069978 KL671190 KFW81622.1 KL273096 KFZ65026.1 KK502193 KFP21496.1 KL224509 KFW60814.1 KK579672 KFW12710.1 AGCU01099544 AGCU01099545 ABQF01060646 ABQF01060647 ABQF01060648 ABQF01060649 ABQF01060650 KK734525 KFR09405.1 KL387282 KFP90344.1 KK432063 KFQ89232.1 KL205953 KFV77530.1 KK844249 KFP86176.1 KK803132 KFQ30242.1 AGTO01020873 KK454255 KFQ75397.1 KK507983 KFP62032.1 MUZQ01000025 OWK62533.1 ABQF01033289 ABQF01033290 KL669021 KFW75147.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000078492
UP000036403
UP000007755
+ More
UP000078541 UP000005205 UP000075809 UP000005203 UP000242457 UP000078542 UP000053097 UP000279307 UP000078540 UP000000311 UP000053105 UP000053825 UP000076858 UP000076502 UP000218220 UP000002358 UP000221080 UP000287033 UP000001555 UP000006813 UP000261440 UP000034805 UP000192224 UP000018468 UP000085678 UP000008672 UP000002279 UP000053286 UP000053858 UP000005447 UP000053283 UP000007648 UP000053258 UP000053119 UP000054081 UP000007267 UP000007754 UP000053605 UP000291020 UP000053584 UP000016665 UP000197619
UP000078541 UP000005205 UP000075809 UP000005203 UP000242457 UP000078542 UP000053097 UP000279307 UP000078540 UP000000311 UP000053105 UP000053825 UP000076858 UP000076502 UP000218220 UP000002358 UP000221080 UP000287033 UP000001555 UP000006813 UP000261440 UP000034805 UP000192224 UP000018468 UP000085678 UP000008672 UP000002279 UP000053286 UP000053858 UP000005447 UP000053283 UP000007648 UP000053258 UP000053119 UP000054081 UP000007267 UP000007754 UP000053605 UP000291020 UP000053584 UP000016665 UP000197619
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JKE9
A0A0N1IEJ1
A0A2H1WII2
A0A194QD05
I4DNY8
A0A2W1BS03
+ More
A0A195EG36 A0A0J7KVT5 F4WQS1 A0A195ESK8 A0A158NRT4 A0A151X3E4 A0A088A7V2 A0A2A3EQV3 A0A195D161 A0A026WCS8 E9IY50 A0A195BRL0 E2A9W5 A0A0N0U5G8 A0A0C9QV25 A0A0L7RBH4 A0A0P4Z5S2 A0A0N8C3W3 A0A0P5G8Y8 A0A0P5BCF9 A0A0P5KH32 A0A0P5KUX9 A0A0P5EJB2 A0A0P5PI74 A0A0P6EJJ2 A0A0P5K871 A0A0P5EQY9 A0A0P5U7G5 A0A023FVG0 A0A1E1XS65 A0A0P6G5E5 A0A0P5GWG1 A0A154PMX7 A0A0P5ZUT9 A0A2A4K2K7 A0A1E1X4T5 A0A131Z7R9 K7J8G8 A0A0P5ENI3 A0A023GKN8 G3MNU7 A0A0N8CT87 A0A0P5K7L3 A0A0P5KQN5 E3TDQ6 A0A131XMH1 A0A2R5LDG3 A0A401RGA2 A0A224Z814 B7Q815 A0A0K8RJW9 Q6NTZ8 G5BZC6 A0A0N8EUU4 Q4PLZ5 A0A0P6JC79 E3TC54 A0A293MXJ0 A0A0P6BV29 A0A3B4EHG4 A0A0P7Y7C1 W5LWG4 A0A2R2MQB0 A0A0P5JR22 H3A0N6 F7E1T8 A0A087R350 A0A0A0ABZ4 A0A286XEB3 A0A091SG92 A0A091VG92 G3WEQ9 A0A093QBY9 A0A094NEQ9 A0A091KCL6 A0A093QQ27 A0A093JCD8 K7F3S2 H0YRE4 A0A091W302 A0A452HA73 A0A091NN40 A0A091UGG0 A0A093HDL5 K7F3S1 A0A091N9R5 A0A091RP19 U3K5A4 A0A091TJV9 A0A091LU97 A0A218V971 H0YXC6 A0A093PSW1
A0A195EG36 A0A0J7KVT5 F4WQS1 A0A195ESK8 A0A158NRT4 A0A151X3E4 A0A088A7V2 A0A2A3EQV3 A0A195D161 A0A026WCS8 E9IY50 A0A195BRL0 E2A9W5 A0A0N0U5G8 A0A0C9QV25 A0A0L7RBH4 A0A0P4Z5S2 A0A0N8C3W3 A0A0P5G8Y8 A0A0P5BCF9 A0A0P5KH32 A0A0P5KUX9 A0A0P5EJB2 A0A0P5PI74 A0A0P6EJJ2 A0A0P5K871 A0A0P5EQY9 A0A0P5U7G5 A0A023FVG0 A0A1E1XS65 A0A0P6G5E5 A0A0P5GWG1 A0A154PMX7 A0A0P5ZUT9 A0A2A4K2K7 A0A1E1X4T5 A0A131Z7R9 K7J8G8 A0A0P5ENI3 A0A023GKN8 G3MNU7 A0A0N8CT87 A0A0P5K7L3 A0A0P5KQN5 E3TDQ6 A0A131XMH1 A0A2R5LDG3 A0A401RGA2 A0A224Z814 B7Q815 A0A0K8RJW9 Q6NTZ8 G5BZC6 A0A0N8EUU4 Q4PLZ5 A0A0P6JC79 E3TC54 A0A293MXJ0 A0A0P6BV29 A0A3B4EHG4 A0A0P7Y7C1 W5LWG4 A0A2R2MQB0 A0A0P5JR22 H3A0N6 F7E1T8 A0A087R350 A0A0A0ABZ4 A0A286XEB3 A0A091SG92 A0A091VG92 G3WEQ9 A0A093QBY9 A0A094NEQ9 A0A091KCL6 A0A093QQ27 A0A093JCD8 K7F3S2 H0YRE4 A0A091W302 A0A452HA73 A0A091NN40 A0A091UGG0 A0A093HDL5 K7F3S1 A0A091N9R5 A0A091RP19 U3K5A4 A0A091TJV9 A0A091LU97 A0A218V971 H0YXC6 A0A093PSW1
PDB
2VYI
E-value=7.54961e-30,
Score=324
Ontologies
GO
Topology
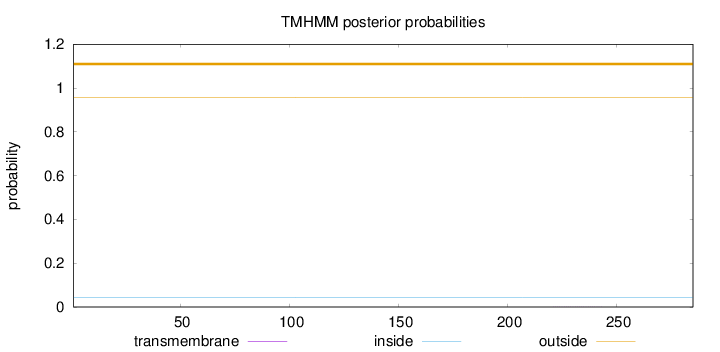
Length:
285
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00512
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.04343
outside
1 - 285
Population Genetic Test Statistics
Pi
269.447697
Theta
218.740862
Tajima's D
0.790129
CLR
0.186721
CSRT
0.596220188990551
Interpretation
Uncertain
