Gene
KWMTBOMO03712
Annotation
PREDICTED:_uncharacterized_protein_LOC105389963_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 1.602
Sequence
CDS
ATGGGTAACTTATCAAAAGAGAGACTTGAAATAGAACCATCGGGGTCAGGTTTTCCATTTTTTAGATGTGGAGTAGATTATGCGGGGCCAATGTTTATACTCAACCGCAAAGGAAGAGGTGCTAAATTAGAAAAATGTTACATTAATTTAATCGTTTGTTTTCTCACTCGCTCGGTATATTTGGATTTGGTCACATCATTAACGACTCAAGCGTACATACTTGCACTTAAAAGGTTCATTTCTAGACGTGGGAAACCATTTCAAATTTTCTCGGACAATGCTAAAACTTTTGTGGGTGCTATGAAGGAATTTTCACAATTCTTGACAGAAAACTCCAAGGAAATTGTCTCGCTAGCAGCCAACGACAACATTAATTTGTCTTTTATACCGCCTTATTCGCCTCATTTTGGTGGTTTATGGGAGGCGGGAGTGAAGTCTTGTAAGCATCACCTACGACGAGTTGTAGGTAATTCACACTTAACGTACGAGGAATTCAGCACGGTATTGGCACAGATAGAAGCCGTCCTGAATTCCCGTCCTATACATCCAATGTCCGCAGATCCAAACGATCTTCGTCCACTCAGCCCTGGTCATTTCCTCATCGGCCGACCGTTGACTGCACCAGCCTGCAGCGACCTCACGGCGACGGCATCACACCGACTGACGCGCTACGATCGAGTGGAACAAATCCGACAACAATTTTGGCAGAGGTGGTCTAAGGAGTACATATCCGAATTGCAGACGAGAACTAAATGGAAAACGGACAAGGAAGACATCGTACCGAACACCCTGGTCCTAATCAAAGAAGACAACCTGCCGCCACTCAAATGGCGTCTTGGACGAGTGCAGCAAACATTTCCTGGCAAGGACGGCATATCCCGAGTAGCTAACCTCAAAACGGCGACTGGGATTGTACAGCGGGCATACTCAAAAATCTGCCCACTCTTACATAATGCAGAGGACTACAACGAGAGTCTCACAACACACACATCGTAA
Protein
MGNLSKERLEIEPSGSGFPFFRCGVDYAGPMFILNRKGRGAKLEKCYINLIVCFLTRSVYLDLVTSLTTQAYILALKRFISRRGKPFQIFSDNAKTFVGAMKEFSQFLTENSKEIVSLAANDNINLSFIPPYSPHFGGLWEAGVKSCKHHLRRVVGNSHLTYEEFSTVLAQIEAVLNSRPIHPMSADPNDLRPLSPGHFLIGRPLTAPACSDLTATASHRLTRYDRVEQIRQQFWQRWSKEYISELQTRTKWKTDKEDIVPNTLVLIKEDNLPPLKWRLGRVQQTFPGKDGISRVANLKTATGIVQRAYSKICPLLHNAEDYNESLTTHTS
Summary
Uniprot
A0A3S2P7Y4
A0A0N0PBK0
S4PWG7
A0A2A4JM97
A0A2A4JBY8
A0A1Y1KBX1
+ More
A0A1Y1MWL6 A0A194RAG7 A0A194RGQ8 A0A1Y1MY22 A0A151K1X2 A0A0J7KLR0 A0A194QJ48 A0A0A9VWY7 A0A0A9WEH4 A0A3S2NVW9 A0A1Y1LN53 A0A0N1I390 A0A151IPF9 A0A0V0GCF2 A0A2L2YR37 A0A1Y1L2Y2 A0A0J7NFF3 A0A0N1IBH6 A0A2S2P0T8 A0A3L8DC69 A0A0J7KE03 D6X1G6 A0A2L2YW58 A0A151JRY2 A0A151I9J7 A0A182GKL8 A0A151JQ29 E9J9Y8 A0A139W8E1 A0A182GDY6 V5FWW0 A0A1Y1L369 A0A0J7K7S0 A0A224XNG8 A0A182GUJ7 A0A0L7QJG1 A0A151J2Q1 A0A1B6EB86 A0A2S2P7L7 A0A194RI04 A0A2W1BII4 A0A2L2YLT6 A0A182G8P0 A0A0J7KDW6 A0A2S2PNZ4 A0A1Y1N6J9 A0A0J7KFV3 A0A151K236 A0A2S2PI44 A0A2L2YNC9 A0A2S2PUJ2 A0A151J2Q3 A0A151JA43 A0A151IGU0 A0A182G7L0 A0A0J7KBV8 A0A0L7QYD7 A0A151ICQ0 A0A0L7RFP1 A0A2A4IUY1 A0A0J7JZS0 A0A0J7JZ44 A0A0J7KD18 A0A1B6CM38 A0A0J7KD57 A0A0A9Z2M8 A0A226CW75 A0A2A4IUL3 E2ARC4 J9K774 A0A1Y1MD76 J9LHB7 D7GYB7 A0A1Y1M979 A0A151IZU4 A0A069DYA1 A0A151IJA6 A0A226CWA5 A0A226DMG9 A0A226EPI5 A0A151IAB8 A0A3L8D4B7 A0A226E226 X1XU48 X1XMQ5 A0A0A9YD88 A0A026WMA2 A0A023EZK8 A0A2S2PTX9 A0A226EUF5 X1XH24 A0A2S2P1P0 A0A023EZB2 J9JT64
A0A1Y1MWL6 A0A194RAG7 A0A194RGQ8 A0A1Y1MY22 A0A151K1X2 A0A0J7KLR0 A0A194QJ48 A0A0A9VWY7 A0A0A9WEH4 A0A3S2NVW9 A0A1Y1LN53 A0A0N1I390 A0A151IPF9 A0A0V0GCF2 A0A2L2YR37 A0A1Y1L2Y2 A0A0J7NFF3 A0A0N1IBH6 A0A2S2P0T8 A0A3L8DC69 A0A0J7KE03 D6X1G6 A0A2L2YW58 A0A151JRY2 A0A151I9J7 A0A182GKL8 A0A151JQ29 E9J9Y8 A0A139W8E1 A0A182GDY6 V5FWW0 A0A1Y1L369 A0A0J7K7S0 A0A224XNG8 A0A182GUJ7 A0A0L7QJG1 A0A151J2Q1 A0A1B6EB86 A0A2S2P7L7 A0A194RI04 A0A2W1BII4 A0A2L2YLT6 A0A182G8P0 A0A0J7KDW6 A0A2S2PNZ4 A0A1Y1N6J9 A0A0J7KFV3 A0A151K236 A0A2S2PI44 A0A2L2YNC9 A0A2S2PUJ2 A0A151J2Q3 A0A151JA43 A0A151IGU0 A0A182G7L0 A0A0J7KBV8 A0A0L7QYD7 A0A151ICQ0 A0A0L7RFP1 A0A2A4IUY1 A0A0J7JZS0 A0A0J7JZ44 A0A0J7KD18 A0A1B6CM38 A0A0J7KD57 A0A0A9Z2M8 A0A226CW75 A0A2A4IUL3 E2ARC4 J9K774 A0A1Y1MD76 J9LHB7 D7GYB7 A0A1Y1M979 A0A151IZU4 A0A069DYA1 A0A151IJA6 A0A226CWA5 A0A226DMG9 A0A226EPI5 A0A151IAB8 A0A3L8D4B7 A0A226E226 X1XU48 X1XMQ5 A0A0A9YD88 A0A026WMA2 A0A023EZK8 A0A2S2PTX9 A0A226EUF5 X1XH24 A0A2S2P1P0 A0A023EZB2 J9JT64
Pubmed
EMBL
RSAL01000222
RVE44041.1
KQ460970
KPJ10166.1
GAIX01006818
JAA85742.1
+ More
NWSH01001014 PCG73111.1 NWSH01001945 PCG69607.1 GEZM01086715 GEZM01086714 JAV59022.1 GEZM01018697 JAV90052.1 KQ460500 KPJ14270.1 KQ460211 KPJ16510.1 GEZM01018702 JAV90048.1 LKEX01009270 KYN50062.1 LBMM01005768 KMQ91222.1 KQ458793 KPJ04955.1 GBHO01042877 JAG00727.1 GBHO01037808 JAG05796.1 RSAL01000052 RVE50155.1 GEZM01052807 JAV74268.1 KQ459256 KPJ02096.1 KQ976863 KYN07859.1 GECL01000456 JAP05668.1 IAAA01044800 LAA09890.1 GEZM01068861 GEZM01068860 JAV66730.1 LBMM01005719 KMQ91270.1 KMQ91271.1 KQ459312 KPJ01687.1 GGMR01010464 MBY23083.1 QOIP01000010 RLU18065.1 LBMM01009062 KMQ88421.1 KQ971371 EFA09479.1 IAAA01066606 LAA12316.1 KQ978591 KYN29960.1 KQ978276 KYM95705.1 JXUM01245428 KQ636870 KXJ62407.1 KQ978749 KYN28625.1 GL769615 EFZ10365.1 KQ973442 KXZ75531.1 JXUM01161881 KQ574228 KXJ67877.1 GALX01006326 JAB62140.1 GEZM01066272 JAV68024.1 LBMM01012392 KMQ86241.1 GFTR01005058 JAW11368.1 JXUM01175059 KQ581415 KXJ62573.1 LHQN01026481 KOC58676.1 KQ980379 KYN16310.1 GEDC01002296 GEDC01002113 JAS35002.1 JAS35185.1 GGMR01012733 MBY25352.1 KQ460205 KPJ16960.1 KZ150101 PZC73504.1 IAAA01034021 IAAA01034022 LAA09083.1 JXUM01152425 KQ571452 KXJ68173.1 LBMM01009098 KMQ88394.1 GGMR01018506 MBY31125.1 GEZM01013857 JAV92285.1 LBMM01008181 KMQ89076.1 LKEX01009172 KYN50051.1 GGMR01016490 MBY29109.1 IAAA01031867 LAA09638.1 GGMR01020500 MBY33119.1 KQ980343 KYN16364.1 KQ979340 KYN21881.1 KQ977667 KYN00648.1 JXUM01150485 KQ571012 KXJ68260.1 LBMM01009941 KMQ87772.1 KQ414692 KOC63602.1 KQ978021 KYM98106.1 KQ414603 KOC69644.1 NWSH01006923 PCG63188.1 LBMM01018894 KMQ83668.1 LBMM01019751 KMQ83463.1 LBMM01009284 KMQ88247.1 GEDC01022769 JAS14529.1 LBMM01009442 KMQ88136.1 GBHO01005508 GBHO01005504 JAG38096.1 JAG38100.1 LNIX01000063 OXA37199.1 PCG63186.1 GL442026 EFN64015.1 ABLF02023723 GEZM01039657 JAV81297.1 ABLF02022040 ABLF02056888 GG695382 EFA13346.1 GEZM01039858 JAV81140.1 KQ980665 KYN14636.1 GBGD01000063 JAC88826.1 KQ977375 KYN03201.1 LNIX01000069 OXA36894.1 LNIX01000016 OXA46198.1 LNIX01000002 OXA59522.1 KQ978265 KYM95834.1 QOIP01000014 RLU14853.1 LNIX01000007 OXA51795.1 ABLF02018371 ABLF02043851 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 KK107156 EZA56791.1 GBBI01004032 JAC14680.1 GGMR01020322 MBY32941.1 OXA60848.1 ABLF02023722 ABLF02057546 GGMR01010750 MBY23369.1 GBBI01004140 JAC14572.1 ABLF02023842 ABLF02028949 ABLF02058391
NWSH01001014 PCG73111.1 NWSH01001945 PCG69607.1 GEZM01086715 GEZM01086714 JAV59022.1 GEZM01018697 JAV90052.1 KQ460500 KPJ14270.1 KQ460211 KPJ16510.1 GEZM01018702 JAV90048.1 LKEX01009270 KYN50062.1 LBMM01005768 KMQ91222.1 KQ458793 KPJ04955.1 GBHO01042877 JAG00727.1 GBHO01037808 JAG05796.1 RSAL01000052 RVE50155.1 GEZM01052807 JAV74268.1 KQ459256 KPJ02096.1 KQ976863 KYN07859.1 GECL01000456 JAP05668.1 IAAA01044800 LAA09890.1 GEZM01068861 GEZM01068860 JAV66730.1 LBMM01005719 KMQ91270.1 KMQ91271.1 KQ459312 KPJ01687.1 GGMR01010464 MBY23083.1 QOIP01000010 RLU18065.1 LBMM01009062 KMQ88421.1 KQ971371 EFA09479.1 IAAA01066606 LAA12316.1 KQ978591 KYN29960.1 KQ978276 KYM95705.1 JXUM01245428 KQ636870 KXJ62407.1 KQ978749 KYN28625.1 GL769615 EFZ10365.1 KQ973442 KXZ75531.1 JXUM01161881 KQ574228 KXJ67877.1 GALX01006326 JAB62140.1 GEZM01066272 JAV68024.1 LBMM01012392 KMQ86241.1 GFTR01005058 JAW11368.1 JXUM01175059 KQ581415 KXJ62573.1 LHQN01026481 KOC58676.1 KQ980379 KYN16310.1 GEDC01002296 GEDC01002113 JAS35002.1 JAS35185.1 GGMR01012733 MBY25352.1 KQ460205 KPJ16960.1 KZ150101 PZC73504.1 IAAA01034021 IAAA01034022 LAA09083.1 JXUM01152425 KQ571452 KXJ68173.1 LBMM01009098 KMQ88394.1 GGMR01018506 MBY31125.1 GEZM01013857 JAV92285.1 LBMM01008181 KMQ89076.1 LKEX01009172 KYN50051.1 GGMR01016490 MBY29109.1 IAAA01031867 LAA09638.1 GGMR01020500 MBY33119.1 KQ980343 KYN16364.1 KQ979340 KYN21881.1 KQ977667 KYN00648.1 JXUM01150485 KQ571012 KXJ68260.1 LBMM01009941 KMQ87772.1 KQ414692 KOC63602.1 KQ978021 KYM98106.1 KQ414603 KOC69644.1 NWSH01006923 PCG63188.1 LBMM01018894 KMQ83668.1 LBMM01019751 KMQ83463.1 LBMM01009284 KMQ88247.1 GEDC01022769 JAS14529.1 LBMM01009442 KMQ88136.1 GBHO01005508 GBHO01005504 JAG38096.1 JAG38100.1 LNIX01000063 OXA37199.1 PCG63186.1 GL442026 EFN64015.1 ABLF02023723 GEZM01039657 JAV81297.1 ABLF02022040 ABLF02056888 GG695382 EFA13346.1 GEZM01039858 JAV81140.1 KQ980665 KYN14636.1 GBGD01000063 JAC88826.1 KQ977375 KYN03201.1 LNIX01000069 OXA36894.1 LNIX01000016 OXA46198.1 LNIX01000002 OXA59522.1 KQ978265 KYM95834.1 QOIP01000014 RLU14853.1 LNIX01000007 OXA51795.1 ABLF02018371 ABLF02043851 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 KK107156 EZA56791.1 GBBI01004032 JAC14680.1 GGMR01020322 MBY32941.1 OXA60848.1 ABLF02023722 ABLF02057546 GGMR01010750 MBY23369.1 GBBI01004140 JAC14572.1 ABLF02023842 ABLF02028949 ABLF02058391
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR001606 ARID_dom
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR008737 Peptidase_asp_put
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR001878 Znf_CCHC
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR001606 ARID_dom
Gene 3D
ProteinModelPortal
A0A3S2P7Y4
A0A0N0PBK0
S4PWG7
A0A2A4JM97
A0A2A4JBY8
A0A1Y1KBX1
+ More
A0A1Y1MWL6 A0A194RAG7 A0A194RGQ8 A0A1Y1MY22 A0A151K1X2 A0A0J7KLR0 A0A194QJ48 A0A0A9VWY7 A0A0A9WEH4 A0A3S2NVW9 A0A1Y1LN53 A0A0N1I390 A0A151IPF9 A0A0V0GCF2 A0A2L2YR37 A0A1Y1L2Y2 A0A0J7NFF3 A0A0N1IBH6 A0A2S2P0T8 A0A3L8DC69 A0A0J7KE03 D6X1G6 A0A2L2YW58 A0A151JRY2 A0A151I9J7 A0A182GKL8 A0A151JQ29 E9J9Y8 A0A139W8E1 A0A182GDY6 V5FWW0 A0A1Y1L369 A0A0J7K7S0 A0A224XNG8 A0A182GUJ7 A0A0L7QJG1 A0A151J2Q1 A0A1B6EB86 A0A2S2P7L7 A0A194RI04 A0A2W1BII4 A0A2L2YLT6 A0A182G8P0 A0A0J7KDW6 A0A2S2PNZ4 A0A1Y1N6J9 A0A0J7KFV3 A0A151K236 A0A2S2PI44 A0A2L2YNC9 A0A2S2PUJ2 A0A151J2Q3 A0A151JA43 A0A151IGU0 A0A182G7L0 A0A0J7KBV8 A0A0L7QYD7 A0A151ICQ0 A0A0L7RFP1 A0A2A4IUY1 A0A0J7JZS0 A0A0J7JZ44 A0A0J7KD18 A0A1B6CM38 A0A0J7KD57 A0A0A9Z2M8 A0A226CW75 A0A2A4IUL3 E2ARC4 J9K774 A0A1Y1MD76 J9LHB7 D7GYB7 A0A1Y1M979 A0A151IZU4 A0A069DYA1 A0A151IJA6 A0A226CWA5 A0A226DMG9 A0A226EPI5 A0A151IAB8 A0A3L8D4B7 A0A226E226 X1XU48 X1XMQ5 A0A0A9YD88 A0A026WMA2 A0A023EZK8 A0A2S2PTX9 A0A226EUF5 X1XH24 A0A2S2P1P0 A0A023EZB2 J9JT64
A0A1Y1MWL6 A0A194RAG7 A0A194RGQ8 A0A1Y1MY22 A0A151K1X2 A0A0J7KLR0 A0A194QJ48 A0A0A9VWY7 A0A0A9WEH4 A0A3S2NVW9 A0A1Y1LN53 A0A0N1I390 A0A151IPF9 A0A0V0GCF2 A0A2L2YR37 A0A1Y1L2Y2 A0A0J7NFF3 A0A0N1IBH6 A0A2S2P0T8 A0A3L8DC69 A0A0J7KE03 D6X1G6 A0A2L2YW58 A0A151JRY2 A0A151I9J7 A0A182GKL8 A0A151JQ29 E9J9Y8 A0A139W8E1 A0A182GDY6 V5FWW0 A0A1Y1L369 A0A0J7K7S0 A0A224XNG8 A0A182GUJ7 A0A0L7QJG1 A0A151J2Q1 A0A1B6EB86 A0A2S2P7L7 A0A194RI04 A0A2W1BII4 A0A2L2YLT6 A0A182G8P0 A0A0J7KDW6 A0A2S2PNZ4 A0A1Y1N6J9 A0A0J7KFV3 A0A151K236 A0A2S2PI44 A0A2L2YNC9 A0A2S2PUJ2 A0A151J2Q3 A0A151JA43 A0A151IGU0 A0A182G7L0 A0A0J7KBV8 A0A0L7QYD7 A0A151ICQ0 A0A0L7RFP1 A0A2A4IUY1 A0A0J7JZS0 A0A0J7JZ44 A0A0J7KD18 A0A1B6CM38 A0A0J7KD57 A0A0A9Z2M8 A0A226CW75 A0A2A4IUL3 E2ARC4 J9K774 A0A1Y1MD76 J9LHB7 D7GYB7 A0A1Y1M979 A0A151IZU4 A0A069DYA1 A0A151IJA6 A0A226CWA5 A0A226DMG9 A0A226EPI5 A0A151IAB8 A0A3L8D4B7 A0A226E226 X1XU48 X1XMQ5 A0A0A9YD88 A0A026WMA2 A0A023EZK8 A0A2S2PTX9 A0A226EUF5 X1XH24 A0A2S2P1P0 A0A023EZB2 J9JT64
Ontologies
GO
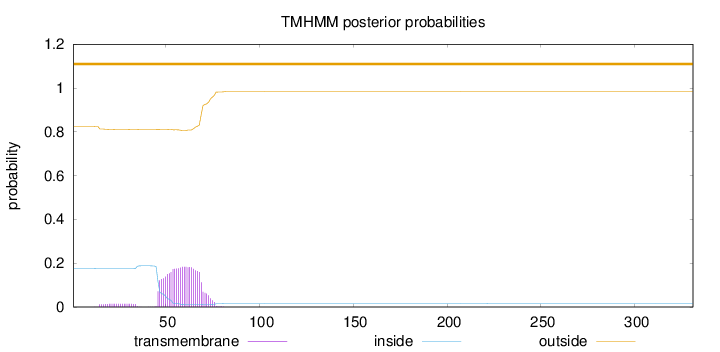
Topology
Length:
331
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.44680999999999
Exp number, first 60 AAs:
2.58506
Total prob of N-in:
0.17517
outside
1 - 331
Population Genetic Test Statistics
Pi
1.581899
Theta
6.534468
Tajima's D
-2.123039
CLR
65.660279
CSRT
0.00674966251687416
Interpretation
Uncertain
