Gene
KWMTBOMO03709 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002747
Annotation
attacin_[Bombyx_mori]
Full name
Attacin
+ More
Attacin-E
Attacin-B
Defense protein 3
Attacin-A
Attacin-E
Attacin-B
Defense protein 3
Attacin-A
Alternative Name
Nuecin
Immune protein P5
Immune protein P5
Location in the cell
Extracellular Reliability : 1.428 Mitochondrial Reliability : 1.707
Sequence
CDS
ATGTCCAAGAGTGTAGCGTTGTTGTTGTTGTGCGCGTGCTTGGCGAGCGGTAGACACGTGCCGACACGTGCGCGGAGACAAGCCGGCAGCTTCACAGTGAACTCGGATGGAACCTCCGGGGCCGCCCTGAAGGTCCCTCTCACCGGCAACGACAAGAACGTACTCAGCGCCATCGGGTCCGCCGACTTCAACGACCGCCACAAGCTCAGCGCCGCCTCCGCAGGGCTCGCTCTGGACAATGTAAACGGGCACGGGCTGAGCCTGACCGGGACCCGCATTCCCGGCTTCGGGGAGCAGCTCGGCGTCGCAGGCAAGGTCAACTTGTTCCACAATAACAACCACGACCTGAGCGCCAAGGCGTTCGCGATCAGGAACTCGCCCAGCGCCATTCCCAACGCGCCCAACTTCAACACGCTGGGCGGCGGAGTGGACTACATGTTCAAACAGAAGGTGGGCGCATCGTTGAGCGCCGCGCACTCTGACGTCATCAACCGGAATGACTACTCGGCCGGGGGTAAGCTGAACCTGTTCCGATCTCCGTCCAGCTCGCTCGACTTCAACGCCGGCTTCAAGAAGTTTGATACGCCTTTTTACAGATCTTCGTGGGAGCCCAATGTGGGATTCTCCTTCTCGAAATTCTTCTAA
Protein
MSKSVALLLLCACLASGRHVPTRARRQAGSFTVNSDGTSGAALKVPLTGNDKNVLSAIGSADFNDRHKLSAASAGLALDNVNGHGLSLTGTRIPGFGEQLGVAGKVNLFHNNNHDLSAKAFAIRNSPSAIPNAPNFNTLGGGVDYMFKQKVGASLSAAHSDVINRNDYSAGGKLNLFRSPSSSLDFNAGFKKFDTPFYRSSWEPNVGFSFSKFF
Summary
Description
Has antibacterial activity against both Gram-positive and Gram-negative bacteria.
Miscellaneous
There are six forms of attacin that are divided into two groups: acidic (E and F) and basic (A, B, C, and D).
Similarity
Belongs to the attacin/sarcotoxin-2 family.
Keywords
Antibiotic
Antimicrobial
Cleavage on pair of basic residues
Complete proteome
Immunity
Innate immunity
Reference proteome
Secreted
Signal
Direct protein sequencing
Repeat
Feature
chain Attacin
Uniprot
Q26431
H9IZR2
D2XRA5
H9IZQ4
J9XS98
Q0VJU9
+ More
Q86M98 P01513 Q56G50 Q95YJ3 P01512 Q95YJ2 B2CZ89 J9XUA5 Q0Q042 Q5MGE6 Q0Q039 Q4PKJ0 P50725 A1E9A9 C5MKE0 A0A0G3DN05 O96361 A0A2H1WMR6 A0A2A4JPN6 A0A2W1BU65 Q58HM7 E2DQE1 A0A2H1WXU6 A0A0G3DJL6 A0A212FKX9 Q0Q041 A0A212FKY7 J9XNB4 A0A0L7LN23 A0A3S2NA53 A0A3S2LMN4 A0A437B4Y1 A0A194PIH0 I4DJ70 D8L125 A0A0N1PI64 A0A437B552 A0A0G3DRX6 A0A437AQV9 A0A212FNR7 A0A194PIR5 I4DJH0 B2CZ90 A0A2H1X0N7 A0A0L7LMR4 A0A437B4Y4 T2C8M9 A0A0N1IEV1 A0A0L7LN30 I4DRF7 A0A212FFC0 A0A1I8NXM0 A0A1I8P7U4 B3NRL8 Q95UA1 Q95NP8 B4HRN6 Q95UA0 P45884 B4QED3 B4QFX9 B4P4I4 Q95U98 A0A0J9RBV8
Q86M98 P01513 Q56G50 Q95YJ3 P01512 Q95YJ2 B2CZ89 J9XUA5 Q0Q042 Q5MGE6 Q0Q039 Q4PKJ0 P50725 A1E9A9 C5MKE0 A0A0G3DN05 O96361 A0A2H1WMR6 A0A2A4JPN6 A0A2W1BU65 Q58HM7 E2DQE1 A0A2H1WXU6 A0A0G3DJL6 A0A212FKX9 Q0Q041 A0A212FKY7 J9XNB4 A0A0L7LN23 A0A3S2NA53 A0A3S2LMN4 A0A437B4Y1 A0A194PIH0 I4DJ70 D8L125 A0A0N1PI64 A0A437B552 A0A0G3DRX6 A0A437AQV9 A0A212FNR7 A0A194PIR5 I4DJH0 B2CZ90 A0A2H1X0N7 A0A0L7LMR4 A0A437B4Y4 T2C8M9 A0A0N1IEV1 A0A0L7LN30 I4DRF7 A0A212FFC0 A0A1I8NXM0 A0A1I8P7U4 B3NRL8 Q95UA1 Q95NP8 B4HRN6 Q95UA0 P45884 B4QED3 B4QFX9 B4P4I4 Q95U98 A0A0J9RBV8
Pubmed
EMBL
S78369
AF005384
D76418
BABH01037979
GU244351
ADB08384.1
+ More
JX294493 AFS30775.1 AM293325 CAL25130.1 AY232304 AAO74640.1 X57715 X00869 M34926 AY960680 AAX58607.1 AB059394 BAB69461.1 X17619 X00787 X62290 M34925 AB059395 BAB69462.1 EU557307 ACB45562.1 JX294494 AFS30776.1 DQ666488 ABG72692.1 AY829840 DQ666491 ABG72695.1 DQ072728 AAY82587.1 U46130 EF114292 ABL63641.1 GQ120502 ACR78454.1 KP056509 AKJ54491.1 AF023277 AAD09288.1 ODYU01009732 SOQ54359.1 NWSH01000918 PCG73544.1 KZ150002 PZC75283.1 AY948540 AAX51192.1 GU111531 ADN51980.1 ODYU01011847 SOQ57822.1 KP056510 AKJ54492.1 AGBW02007944 OWR54405.1 DQ666489 ABG72693.1 OWR54404.1 JX294492 AFS30774.1 JTDY01000509 KOB76847.1 RSAL01000157 RVE45569.1 RSAL01002229 RVE40547.1 RVE45566.1 KQ459602 KPI93211.1 AK401338 BAM17960.1 FJ438392 ACR82290.1 KQ460427 KPJ14761.1 RSAL01000126 RVE45568.1 RVE46563.1 KP056511 AKJ54493.1 RSAL01005472 RVE40007.1 AGBW02004511 OWR55396.1 KPI93212.1 AK401438 BAM18060.1 EU557308 ACB45563.1 ODYU01012449 SOQ58756.1 KOB76848.1 RVE45567.1 KF146178 AGV28582.1 KPJ14762.1 JTDY01000537 KOB76749.1 AK405098 BAM20497.1 AGBW02008836 OWR52427.1 CH954179 EDV56170.2 AY056855 AAL23641.1 AY056857 AY056861 AY056862 AY056863 AY056864 AY056865 AAL23643.1 AAL23646.1 AAL23647.1 AAL23648.1 AAL23649.1 AAL23650.1 CH480816 EDW47900.1 AY056856 AAL23642.1 Z46893 AF220544 AE013599 CM000362 EDX06923.1 CM002911 EDX07124.1 KMY93826.1 CM000158 EDW90623.2 AY056860 AAL23645.1 KMY93476.1
JX294493 AFS30775.1 AM293325 CAL25130.1 AY232304 AAO74640.1 X57715 X00869 M34926 AY960680 AAX58607.1 AB059394 BAB69461.1 X17619 X00787 X62290 M34925 AB059395 BAB69462.1 EU557307 ACB45562.1 JX294494 AFS30776.1 DQ666488 ABG72692.1 AY829840 DQ666491 ABG72695.1 DQ072728 AAY82587.1 U46130 EF114292 ABL63641.1 GQ120502 ACR78454.1 KP056509 AKJ54491.1 AF023277 AAD09288.1 ODYU01009732 SOQ54359.1 NWSH01000918 PCG73544.1 KZ150002 PZC75283.1 AY948540 AAX51192.1 GU111531 ADN51980.1 ODYU01011847 SOQ57822.1 KP056510 AKJ54492.1 AGBW02007944 OWR54405.1 DQ666489 ABG72693.1 OWR54404.1 JX294492 AFS30774.1 JTDY01000509 KOB76847.1 RSAL01000157 RVE45569.1 RSAL01002229 RVE40547.1 RVE45566.1 KQ459602 KPI93211.1 AK401338 BAM17960.1 FJ438392 ACR82290.1 KQ460427 KPJ14761.1 RSAL01000126 RVE45568.1 RVE46563.1 KP056511 AKJ54493.1 RSAL01005472 RVE40007.1 AGBW02004511 OWR55396.1 KPI93212.1 AK401438 BAM18060.1 EU557308 ACB45563.1 ODYU01012449 SOQ58756.1 KOB76848.1 RVE45567.1 KF146178 AGV28582.1 KPJ14762.1 JTDY01000537 KOB76749.1 AK405098 BAM20497.1 AGBW02008836 OWR52427.1 CH954179 EDV56170.2 AY056855 AAL23641.1 AY056857 AY056861 AY056862 AY056863 AY056864 AY056865 AAL23643.1 AAL23646.1 AAL23647.1 AAL23648.1 AAL23649.1 AAL23650.1 CH480816 EDW47900.1 AY056856 AAL23642.1 Z46893 AF220544 AE013599 CM000362 EDX06923.1 CM002911 EDX07124.1 KMY93826.1 CM000158 EDW90623.2 AY056860 AAL23645.1 KMY93476.1
Proteomes
Interpro
ProteinModelPortal
Q26431
H9IZR2
D2XRA5
H9IZQ4
J9XS98
Q0VJU9
+ More
Q86M98 P01513 Q56G50 Q95YJ3 P01512 Q95YJ2 B2CZ89 J9XUA5 Q0Q042 Q5MGE6 Q0Q039 Q4PKJ0 P50725 A1E9A9 C5MKE0 A0A0G3DN05 O96361 A0A2H1WMR6 A0A2A4JPN6 A0A2W1BU65 Q58HM7 E2DQE1 A0A2H1WXU6 A0A0G3DJL6 A0A212FKX9 Q0Q041 A0A212FKY7 J9XNB4 A0A0L7LN23 A0A3S2NA53 A0A3S2LMN4 A0A437B4Y1 A0A194PIH0 I4DJ70 D8L125 A0A0N1PI64 A0A437B552 A0A0G3DRX6 A0A437AQV9 A0A212FNR7 A0A194PIR5 I4DJH0 B2CZ90 A0A2H1X0N7 A0A0L7LMR4 A0A437B4Y4 T2C8M9 A0A0N1IEV1 A0A0L7LN30 I4DRF7 A0A212FFC0 A0A1I8NXM0 A0A1I8P7U4 B3NRL8 Q95UA1 Q95NP8 B4HRN6 Q95UA0 P45884 B4QED3 B4QFX9 B4P4I4 Q95U98 A0A0J9RBV8
Q86M98 P01513 Q56G50 Q95YJ3 P01512 Q95YJ2 B2CZ89 J9XUA5 Q0Q042 Q5MGE6 Q0Q039 Q4PKJ0 P50725 A1E9A9 C5MKE0 A0A0G3DN05 O96361 A0A2H1WMR6 A0A2A4JPN6 A0A2W1BU65 Q58HM7 E2DQE1 A0A2H1WXU6 A0A0G3DJL6 A0A212FKX9 Q0Q041 A0A212FKY7 J9XNB4 A0A0L7LN23 A0A3S2NA53 A0A3S2LMN4 A0A437B4Y1 A0A194PIH0 I4DJ70 D8L125 A0A0N1PI64 A0A437B552 A0A0G3DRX6 A0A437AQV9 A0A212FNR7 A0A194PIR5 I4DJH0 B2CZ90 A0A2H1X0N7 A0A0L7LMR4 A0A437B4Y4 T2C8M9 A0A0N1IEV1 A0A0L7LN30 I4DRF7 A0A212FFC0 A0A1I8NXM0 A0A1I8P7U4 B3NRL8 Q95UA1 Q95NP8 B4HRN6 Q95UA0 P45884 B4QED3 B4QFX9 B4P4I4 Q95U98 A0A0J9RBV8
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 17,
Likelihood: 0.938825
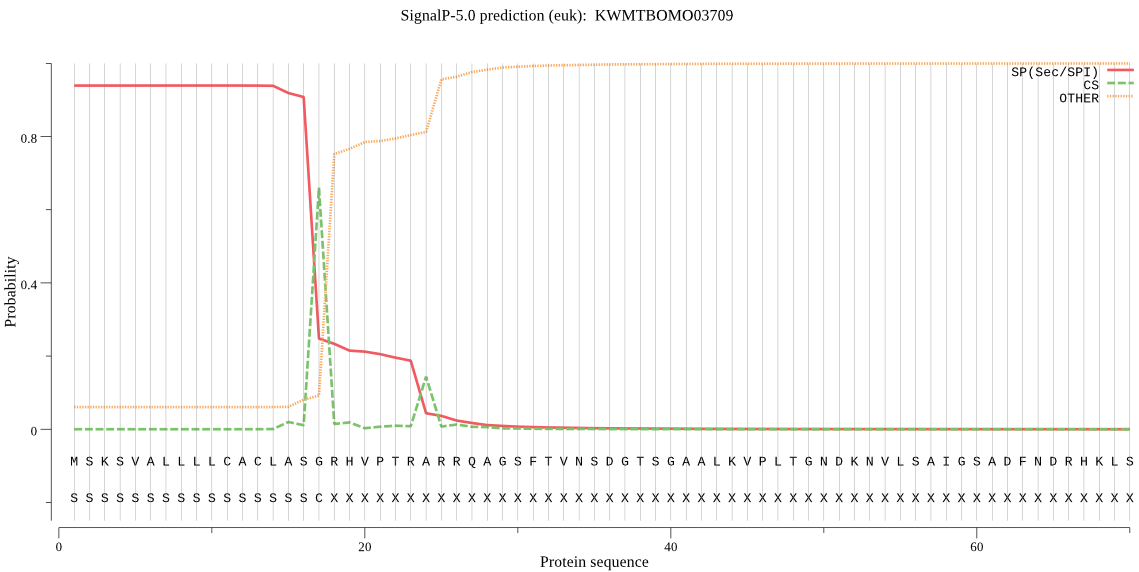
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.265350000000001
Exp number, first 60 AAs:
0.26453
Total prob of N-in:
0.02044
outside
1 - 214

Population Genetic Test Statistics
Pi
27.671084
Theta
21.047349
Tajima's D
0.8305
CLR
0.206678
CSRT
0.607519624018799
Interpretation
Uncertain
