Gene
KWMTBOMO03705
Pre Gene Modal
BGIBMGA002735
Annotation
neuropeptide_FF_receptor_2-like_[Bombyx_mori]
Full name
Neuropeptide SIFamide receptor
Location in the cell
PlasmaMembrane Reliability : 2.911
Sequence
CDS
ATGGAACGTATGCAGCAAAAATCCAAGGTCAAAGTCGTCAAAATGCTGGTTGCTGTTGTGATCCTGTTTGTTCTTTCCTGGTTTCCATTGTACCTGATCTTTGCAAGGATAAAACTGGGAGGACCGATCAAGAAATGGGAGGAAGAGATGCTTCCGATCGTGACACCACTGGCTCAATGGCTTGGAGCCTCGAACTCTTGCATCAACCCGATCCTGTACGCGTTTTTCAACAAGAAGTACAGAAAGGGATTTGTAGCCATCATCAAGAGCAGGAAATGCTGCGGCCGACTCCGCTACTACGAGACCATTGCTCTGCAGTCCTCGAGCACTTCCACCAGAAAGTCTTGGCATTACAACAACAATAACCCGTCGATCACCCGGCGGTCCCCACCGGTCGACAAAAACGCCGTGTCTTTTATTTTCAGTCACACAGGCGTGTAA
Protein
MERMQQKSKVKVVKMLVAVVILFVLSWFPLYLIFARIKLGGPIKKWEEEMLPIVTPLAQWLGASNSCINPILYAFFNKKYRKGFVAIIKSRKCCGRLRYYETIALQSSSTSTRKSWHYNNNNPSITRRSPPVDKNAVSFIFSHTGV
Summary
Description
Receptor for the neuropeptide SIFamide (PubMed:16378592). Modulates sexual behavior by playing a role in male sex discrimination (PubMed:26469541). Also involved in promoting sleep (PubMed:24658384).
Miscellaneous
The sequence shown here is derived from an EMBL/GenBank/DDBJ third party annotation (TPA) entry.
Similarity
Belongs to the G-protein coupled receptor 1 family.
Keywords
Behavior
Cell membrane
Complete proteome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Reference proteome
Transducer
Transmembrane
Transmembrane helix
Feature
chain Neuropeptide SIFamide receptor
Uniprot
H9IZQ0
J7RNN2
A0A2A4JRE5
A0A212FF40
A0A2H1WUE5
A0A2W1BST6
+ More
A0A194PX61 A0A0S1YDQ5 A0A0L7LVD5 A0A194R8R7 A0A0T6BA96 A0A2P8ZLN2 A0A139WAW3 N6TX07 D6X402 A0A1W4X1F3 K7J8L5 A0A232EF77 A0A2J7QJK6 A0A1B0BYC3 A0A1B0G654 A0A1A9Z9I0 A0A1A9YLN7 A0A1I8NLV7 A0A1I8NLV5 A0A1I8MRR4 A0A1B0CT13 A0A0L0CGZ4 A0A182TBN2 A0A154P271 A0A411JKH9 A0A336LUR7 A0A182HDD7 A0A1J1INC1 B0XDM7 J9HIW7 U3UA00 W8C2A8 A0A182NFU2 A0A182VXR9 A0A182P8P8 A0A182M089 Q16V26 J7SF97 A0A1S4FN27 A0A182YRL1 A0A182KWQ4 A0A182JK22 A0A182FP85 A0A0J7L5V7 A0A084VC27 A0A182RQQ7 A0A182U2Y2 A0A182V1E9 W5JQJ2 A0A182HJJ9 A0A182XKE5 A0A182QPX0 B4PQD9 Q7Q9B7 A0A3B0K4K1 A0A067RKW7 E2C2H8 A0A2S2NDF6 B3LZ35 A0A1B6HC70 Q8IN35 B4IKH5 Q29B01 A0A1W4VYF2 B3P029 B4GZ96 A0A3L8DP57 A0A2H8TNJ5 B4R117 J9K651 A0A2S2R4C1 A0A151IQP4 E9J618 B4MBG4 F4X6G7 A0A195F227 A0A158NFZ3 A0A0M4EWH8 A0A195B2S7 A0A195E743 A0A151XC59 B4NKH1 A0A0L7QLJ1 B4KCU4 B4JS44 E2AMG1 A0A310SPZ9 E0VGM1 A0A2R7WSQ9 A0A088A708 A0A2A3E6N8 A9X454 A0A0N0BCH8
A0A194PX61 A0A0S1YDQ5 A0A0L7LVD5 A0A194R8R7 A0A0T6BA96 A0A2P8ZLN2 A0A139WAW3 N6TX07 D6X402 A0A1W4X1F3 K7J8L5 A0A232EF77 A0A2J7QJK6 A0A1B0BYC3 A0A1B0G654 A0A1A9Z9I0 A0A1A9YLN7 A0A1I8NLV7 A0A1I8NLV5 A0A1I8MRR4 A0A1B0CT13 A0A0L0CGZ4 A0A182TBN2 A0A154P271 A0A411JKH9 A0A336LUR7 A0A182HDD7 A0A1J1INC1 B0XDM7 J9HIW7 U3UA00 W8C2A8 A0A182NFU2 A0A182VXR9 A0A182P8P8 A0A182M089 Q16V26 J7SF97 A0A1S4FN27 A0A182YRL1 A0A182KWQ4 A0A182JK22 A0A182FP85 A0A0J7L5V7 A0A084VC27 A0A182RQQ7 A0A182U2Y2 A0A182V1E9 W5JQJ2 A0A182HJJ9 A0A182XKE5 A0A182QPX0 B4PQD9 Q7Q9B7 A0A3B0K4K1 A0A067RKW7 E2C2H8 A0A2S2NDF6 B3LZ35 A0A1B6HC70 Q8IN35 B4IKH5 Q29B01 A0A1W4VYF2 B3P029 B4GZ96 A0A3L8DP57 A0A2H8TNJ5 B4R117 J9K651 A0A2S2R4C1 A0A151IQP4 E9J618 B4MBG4 F4X6G7 A0A195F227 A0A158NFZ3 A0A0M4EWH8 A0A195B2S7 A0A195E743 A0A151XC59 B4NKH1 A0A0L7QLJ1 B4KCU4 B4JS44 E2AMG1 A0A310SPZ9 E0VGM1 A0A2R7WSQ9 A0A088A708 A0A2A3E6N8 A9X454 A0A0N0BCH8
Pubmed
19121390
16378592
22118469
28756777
26354079
26227816
+ More
29403074 18362917 19820115 23537049 18054377 18025266 20075255 28648823 25315136 26108605 30735632 26483478 17510324 23932938 24495485 25244985 20966253 24438588 20920257 23761445 17994087 17550304 12364791 14747013 17210077 24845553 20798317 10731132 12537572 24658384 26469541 15632085 30249741 21282665 21719571 21347285 18057021 20566863
29403074 18362917 19820115 23537049 18054377 18025266 20075255 28648823 25315136 26108605 30735632 26483478 17510324 23932938 24495485 25244985 20966253 24438588 20920257 23761445 17994087 17550304 12364791 14747013 17210077 24845553 20798317 10731132 12537572 24658384 26469541 15632085 30249741 21282665 21719571 21347285 18057021 20566863
EMBL
BABH01038005
BK005732
DAA35192.1
NWSH01000728
PCG74561.1
AGBW02008857
+ More
OWR52361.1 ODYU01011090 SOQ56612.1 KZ149985 PZC75706.1 KQ459589 KPI97603.1 KT031041 RSAL01000004 ALM88339.1 RVE54563.1 JTDY01000013 KOB79427.1 KQ460500 KPJ14233.1 LJIG01002682 KRT84258.1 PYGN01000021 PSN57408.1 KQ971379 KYB25074.1 APGK01048267 KB741103 KB632401 ENN73845.1 ERL94912.1 BK005734 DAA35194.1 EEZ97714.1 AAZX01001552 NNAY01005093 OXU17010.1 NEVH01013552 PNF28767.1 JXJN01022584 CCAG010005914 AJWK01026873 JRES01000409 KNC31511.1 KQ434803 KZC05971.1 MH810199 QBC65464.1 UFQT01000121 SSX20423.1 JXUM01000331 JXUM01000332 JXUM01000333 KQ560105 KXJ84523.1 CVRI01000055 CRL01052.1 DS232769 EDS45535.1 CH477605 EJY57824.1 AB817286 BAO01053.1 GAMC01010686 GAMC01010685 JAB95869.1 AXCM01001598 EAT38404.2 BK005733 DAA35193.1 LBMM01000634 KMQ97938.1 ATLV01009984 ATLV01009985 KE524560 KFB35521.1 ADMH02000799 ETN65014.1 APCN01002178 AXCN02000246 CM000160 EDW96248.2 AAAB01008904 EAA09601.4 OUUW01000013 SPP88223.1 KK852410 KDR24477.1 GL452135 EFN77834.1 GGMR01002614 MBY15233.1 CH902617 EDV41909.1 GECU01035438 JAS72268.1 DQ285411 AE014297 BT009988 AAQ22457.1 ABB96223.1 CH480853 EDW51579.1 CM000070 EAL27198.4 CH954181 EDV48265.1 CH479198 EDW28114.1 QOIP01000006 RLU22214.1 GFXV01003686 MBW15491.1 CM000364 EDX12104.1 ABLF02029597 ABLF02029598 GGMS01015653 MBY84856.1 KQ976780 KYN08363.1 GL768221 EFZ11709.1 CH940656 EDW58435.2 GL888818 EGI57927.1 KQ981864 KYN34222.1 ADTU01014861 ADTU01014862 ADTU01014863 CP012526 ALC47905.1 KQ976662 KYM78585.1 KQ979568 KYN20916.1 KQ982314 KYQ57878.1 CH964272 EDW85143.2 KQ414915 KOC59498.1 CH933806 EDW16966.2 KRG02418.1 KRG02419.1 CH916373 EDV94584.1 GL440813 EFN65385.1 KQ760287 OAD60890.1 DS235150 EEB12527.1 KK855359 PTY22170.1 KZ288357 PBC27164.1 DQ339143 ABC68306.1 KQ435908 KOX69018.1
OWR52361.1 ODYU01011090 SOQ56612.1 KZ149985 PZC75706.1 KQ459589 KPI97603.1 KT031041 RSAL01000004 ALM88339.1 RVE54563.1 JTDY01000013 KOB79427.1 KQ460500 KPJ14233.1 LJIG01002682 KRT84258.1 PYGN01000021 PSN57408.1 KQ971379 KYB25074.1 APGK01048267 KB741103 KB632401 ENN73845.1 ERL94912.1 BK005734 DAA35194.1 EEZ97714.1 AAZX01001552 NNAY01005093 OXU17010.1 NEVH01013552 PNF28767.1 JXJN01022584 CCAG010005914 AJWK01026873 JRES01000409 KNC31511.1 KQ434803 KZC05971.1 MH810199 QBC65464.1 UFQT01000121 SSX20423.1 JXUM01000331 JXUM01000332 JXUM01000333 KQ560105 KXJ84523.1 CVRI01000055 CRL01052.1 DS232769 EDS45535.1 CH477605 EJY57824.1 AB817286 BAO01053.1 GAMC01010686 GAMC01010685 JAB95869.1 AXCM01001598 EAT38404.2 BK005733 DAA35193.1 LBMM01000634 KMQ97938.1 ATLV01009984 ATLV01009985 KE524560 KFB35521.1 ADMH02000799 ETN65014.1 APCN01002178 AXCN02000246 CM000160 EDW96248.2 AAAB01008904 EAA09601.4 OUUW01000013 SPP88223.1 KK852410 KDR24477.1 GL452135 EFN77834.1 GGMR01002614 MBY15233.1 CH902617 EDV41909.1 GECU01035438 JAS72268.1 DQ285411 AE014297 BT009988 AAQ22457.1 ABB96223.1 CH480853 EDW51579.1 CM000070 EAL27198.4 CH954181 EDV48265.1 CH479198 EDW28114.1 QOIP01000006 RLU22214.1 GFXV01003686 MBW15491.1 CM000364 EDX12104.1 ABLF02029597 ABLF02029598 GGMS01015653 MBY84856.1 KQ976780 KYN08363.1 GL768221 EFZ11709.1 CH940656 EDW58435.2 GL888818 EGI57927.1 KQ981864 KYN34222.1 ADTU01014861 ADTU01014862 ADTU01014863 CP012526 ALC47905.1 KQ976662 KYM78585.1 KQ979568 KYN20916.1 KQ982314 KYQ57878.1 CH964272 EDW85143.2 KQ414915 KOC59498.1 CH933806 EDW16966.2 KRG02418.1 KRG02419.1 CH916373 EDV94584.1 GL440813 EFN65385.1 KQ760287 OAD60890.1 DS235150 EEB12527.1 KK855359 PTY22170.1 KZ288357 PBC27164.1 DQ339143 ABC68306.1 KQ435908 KOX69018.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000037510
+ More
UP000053240 UP000245037 UP000007266 UP000019118 UP000030742 UP000192223 UP000002358 UP000215335 UP000235965 UP000092460 UP000092444 UP000092445 UP000092443 UP000095300 UP000095301 UP000092461 UP000037069 UP000075901 UP000076502 UP000069940 UP000249989 UP000183832 UP000002320 UP000008820 UP000075884 UP000075920 UP000075885 UP000075883 UP000076408 UP000075882 UP000075880 UP000069272 UP000036403 UP000030765 UP000075900 UP000075902 UP000075903 UP000000673 UP000075840 UP000076407 UP000075886 UP000002282 UP000007062 UP000268350 UP000027135 UP000008237 UP000007801 UP000000803 UP000001292 UP000001819 UP000192221 UP000008711 UP000008744 UP000279307 UP000000304 UP000007819 UP000078542 UP000008792 UP000007755 UP000078541 UP000005205 UP000092553 UP000078540 UP000078492 UP000075809 UP000007798 UP000053825 UP000009192 UP000001070 UP000000311 UP000009046 UP000005203 UP000242457 UP000053105
UP000053240 UP000245037 UP000007266 UP000019118 UP000030742 UP000192223 UP000002358 UP000215335 UP000235965 UP000092460 UP000092444 UP000092445 UP000092443 UP000095300 UP000095301 UP000092461 UP000037069 UP000075901 UP000076502 UP000069940 UP000249989 UP000183832 UP000002320 UP000008820 UP000075884 UP000075920 UP000075885 UP000075883 UP000076408 UP000075882 UP000075880 UP000069272 UP000036403 UP000030765 UP000075900 UP000075902 UP000075903 UP000000673 UP000075840 UP000076407 UP000075886 UP000002282 UP000007062 UP000268350 UP000027135 UP000008237 UP000007801 UP000000803 UP000001292 UP000001819 UP000192221 UP000008711 UP000008744 UP000279307 UP000000304 UP000007819 UP000078542 UP000008792 UP000007755 UP000078541 UP000005205 UP000092553 UP000078540 UP000078492 UP000075809 UP000007798 UP000053825 UP000009192 UP000001070 UP000000311 UP000009046 UP000005203 UP000242457 UP000053105
Interpro
ProteinModelPortal
H9IZQ0
J7RNN2
A0A2A4JRE5
A0A212FF40
A0A2H1WUE5
A0A2W1BST6
+ More
A0A194PX61 A0A0S1YDQ5 A0A0L7LVD5 A0A194R8R7 A0A0T6BA96 A0A2P8ZLN2 A0A139WAW3 N6TX07 D6X402 A0A1W4X1F3 K7J8L5 A0A232EF77 A0A2J7QJK6 A0A1B0BYC3 A0A1B0G654 A0A1A9Z9I0 A0A1A9YLN7 A0A1I8NLV7 A0A1I8NLV5 A0A1I8MRR4 A0A1B0CT13 A0A0L0CGZ4 A0A182TBN2 A0A154P271 A0A411JKH9 A0A336LUR7 A0A182HDD7 A0A1J1INC1 B0XDM7 J9HIW7 U3UA00 W8C2A8 A0A182NFU2 A0A182VXR9 A0A182P8P8 A0A182M089 Q16V26 J7SF97 A0A1S4FN27 A0A182YRL1 A0A182KWQ4 A0A182JK22 A0A182FP85 A0A0J7L5V7 A0A084VC27 A0A182RQQ7 A0A182U2Y2 A0A182V1E9 W5JQJ2 A0A182HJJ9 A0A182XKE5 A0A182QPX0 B4PQD9 Q7Q9B7 A0A3B0K4K1 A0A067RKW7 E2C2H8 A0A2S2NDF6 B3LZ35 A0A1B6HC70 Q8IN35 B4IKH5 Q29B01 A0A1W4VYF2 B3P029 B4GZ96 A0A3L8DP57 A0A2H8TNJ5 B4R117 J9K651 A0A2S2R4C1 A0A151IQP4 E9J618 B4MBG4 F4X6G7 A0A195F227 A0A158NFZ3 A0A0M4EWH8 A0A195B2S7 A0A195E743 A0A151XC59 B4NKH1 A0A0L7QLJ1 B4KCU4 B4JS44 E2AMG1 A0A310SPZ9 E0VGM1 A0A2R7WSQ9 A0A088A708 A0A2A3E6N8 A9X454 A0A0N0BCH8
A0A194PX61 A0A0S1YDQ5 A0A0L7LVD5 A0A194R8R7 A0A0T6BA96 A0A2P8ZLN2 A0A139WAW3 N6TX07 D6X402 A0A1W4X1F3 K7J8L5 A0A232EF77 A0A2J7QJK6 A0A1B0BYC3 A0A1B0G654 A0A1A9Z9I0 A0A1A9YLN7 A0A1I8NLV7 A0A1I8NLV5 A0A1I8MRR4 A0A1B0CT13 A0A0L0CGZ4 A0A182TBN2 A0A154P271 A0A411JKH9 A0A336LUR7 A0A182HDD7 A0A1J1INC1 B0XDM7 J9HIW7 U3UA00 W8C2A8 A0A182NFU2 A0A182VXR9 A0A182P8P8 A0A182M089 Q16V26 J7SF97 A0A1S4FN27 A0A182YRL1 A0A182KWQ4 A0A182JK22 A0A182FP85 A0A0J7L5V7 A0A084VC27 A0A182RQQ7 A0A182U2Y2 A0A182V1E9 W5JQJ2 A0A182HJJ9 A0A182XKE5 A0A182QPX0 B4PQD9 Q7Q9B7 A0A3B0K4K1 A0A067RKW7 E2C2H8 A0A2S2NDF6 B3LZ35 A0A1B6HC70 Q8IN35 B4IKH5 Q29B01 A0A1W4VYF2 B3P029 B4GZ96 A0A3L8DP57 A0A2H8TNJ5 B4R117 J9K651 A0A2S2R4C1 A0A151IQP4 E9J618 B4MBG4 F4X6G7 A0A195F227 A0A158NFZ3 A0A0M4EWH8 A0A195B2S7 A0A195E743 A0A151XC59 B4NKH1 A0A0L7QLJ1 B4KCU4 B4JS44 E2AMG1 A0A310SPZ9 E0VGM1 A0A2R7WSQ9 A0A088A708 A0A2A3E6N8 A9X454 A0A0N0BCH8
PDB
3PBL
E-value=0.000465654,
Score=97
Ontologies
KEGG
GO
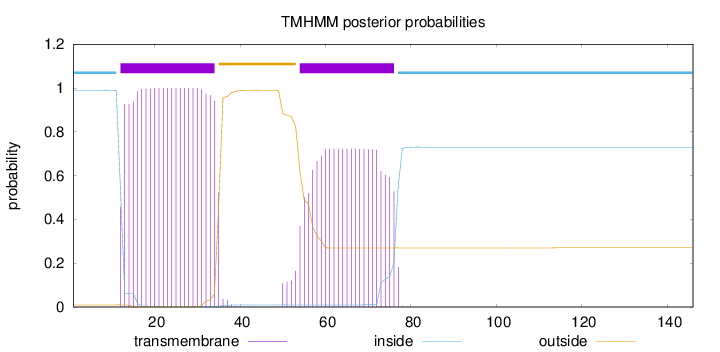
Topology
Subcellular location
Cell membrane
Length:
146
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.50213
Exp number, first 60 AAs:
27.31438
Total prob of N-in:
0.99038
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 53
TMhelix
54 - 76
inside
77 - 146
Population Genetic Test Statistics
Pi
21.715816
Theta
18.029274
Tajima's D
0.619475
CLR
0
CSRT
0.55757212139393
Interpretation
Uncertain
